Day 4, in anticipation of World DNA day this Sunday, today we want to shine a light on how genetic analysis can be done today, and support disease diagnosis, treatment, and prevention.
In the evolution of genetic testing, as described in “What is DNA and what is its size?“, the current next-generation sequencing (NGS) methodologies have substantially increased the sequencing yield and have lowered the cost per base [1]. This development now allows a highly faster turnaround time in genetic diagnostics, but what are the steps from the initial consultation to receiving the genetic report?
Consultation & Sample Collection
Prior to the diagnostic testing, an initial genetic consultation aims to provide the patient with all the support and information as needed to understand the process and make an informed decision on genetic analysis. Not every consultation leads ultimately to genetic diagnostics. Nowadays, the information provided by genetic professionals can help the patient and even their families in understanding the medical, psychological, and familial implications of genetic disease-related concerns, and making the decision.
Should genetic testing prove helpful in the context of disease diagnosis, prevention or for the selection of appropriate treatment, sample collection will follow. Usually, the DNA is extracted from a blood sample, but genetic material can also be obtained from a saliva sample.
Sequencing
This sample is prepared in the laboratory and then sequenced using suitable sequencing machines. Since 2010 the NGS technology has substituted mostly the previous traditional standard of Sanger sequencing developed in the late 1970s. The conventional approach only focused on single genes to be sequenced and solely one individual’s DNA at a time[2]. Thanks to NGS technology, it became routinely available the possibility to analyze multiple individuals in parallel for a set of genes with what are called gene panels. Thanks to the continuous progress of NGS technology and the piling-up of the resulting knowledge, we can analyze the entire genome today with a high degree of automation [3].
The result of the sequencing process is a file containing the individual’s sequence of the bases of their DNA, for the area of interest for the particular diagnostic question.
More information on sequencing you can find here.
Analysis
During the analysis, a complex series of bioinformatic processes are carried over in order to align the sequence obtained from the individual to a reference genome, and to determine variations in the DNA sequence of the individual. The so-called genetic variants can involve single or multiple bases, and in some cases disrupt specific biological functions turning out as disease-causing [4]. After the identification of genetic variants, an expert provides an interpretation availing of a wide set of resources or annotation databases for genetic data. The knowledge in these databases can provide information on the role that identified variants might play in specific diseases, and finally lead to a report with supporting information for further medical action.
The knowledge associated to genetic variation is constantly growing, and it is a lengthy effort by the entire research community worldwide to obtain reliable data to base medical decisions regarding prevention or treatment.
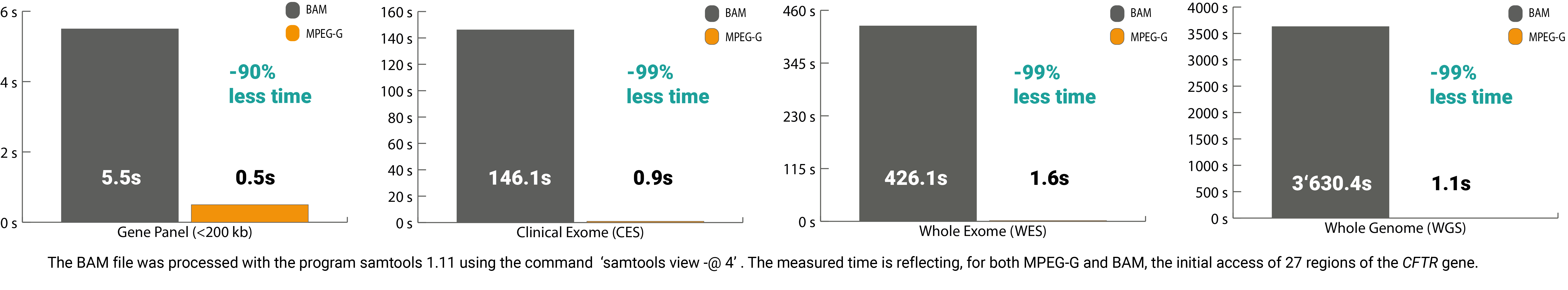
Over the last decade, the efficiency along this process chain for genetic diagnostics has increased significantly. The sequencing step has become a highly automated process, directly impacting the subsequent analysis. The processing of this massive amount of electronic data can be very time-consuming. Currently, solely the digital preprocessing and accessing of regions on the DNA sequence in legacy formats – especially when analyzing whole-exome and whole-genome sequencing datasets – can take up to multiple hours. Resulting in a delay for the start of the analysis, interpretation, and ultimately slowing down the delivery of the needed report for the patient.
The ISO/IEC 23092 standard series (MPEG-G) published by ISO is ideal for improving time efficiency along the processing chain for genome sequencing data. This open international standard is structured so that it leads to a minimal time delay until data consumption. Thereby this data format contains so-called access units that can be opened and processed simultaneously. Compared to legacy formats, which can only process data sequentially, MPEG-G shortens the processing time considerably.
In addition, the selective access feature of the standard also delivers benefits in terms of time. The core lies in the indexing structure of MPEG-G, which allows quick access to the desired offset without spending time sorting and indexing, resulting in massive time savings. For example, without the pre-processing steps, the processing time to access 27 regions of interest in the CFTR gene from a whole-exome sequencing file, only 2secs vs. 426secs with legacy formats.
Especially for laboratories, GenomSys offers a specific solution based on the ISO/IEC 23092 standard. GenomSys Variant Analyzer (GVA), a platform natively operating on MPEG-G genomic data format that enables accurate variants identification, annotation, and interpretation, the genetic professional in the laboratory can benefit directly from this faster processing time.
By Lucas Laner on April 23, 2021.
References:
[1] Buermans HPJ, den Dunnen JT. Next generation sequencing technology: Advances and applications. Biochimica et Biophysica Acta (BBA) – Molecular Basis of Disease; From genome to function 2014 10/01;1842(10):1932-1941.[2] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3917434/
[3] https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Fact-Sheet
[4] Trotta L.. Genetics of Primary Immunodeficiency in Finland https://helda.helsinki.fi/bitstream/handle/10138/278894/TROTTA_e-thesis_19122018.pdf?sequence=1&isAllowed=y





