Following our last week’s article of the series “MPEG-G, can I eat it?” explaining MPEG-G and its history, today’s article is supposed to shed light on one of the four main benefits of the MPEG-G standard – the compression rate for genomic datasets. We want to explain why genomic data is in need of higher file compression, how the MPEG-G compression benefit works, and give a practical example for a laboratory handling genomic data.
In case you missed our last week’s article, you can find it here:
MPEG-G, can I eat it? Definition & History
More data, more problems?
Like any other industry, Genomic testing has received clear benefits from technological advancements, but challenges also accompany these. The advent of new sequencing technology, the possibility of analyzing larger DNA areas, and the general digitalization were critical drivers for these new challenges.
Over the past decade, the world has been experiencing a drastic increase in data generation. In 2013 the global data volume was 3.5 Zettabytes; 1 Zettabytes is equal to 1 trillion Gigabytes just to put it into perspective.[1] By 2018, it had already reached 33 Zettabytes, and it is estimated to climb up to 175 Zettabytes by 2025 This thirst for data and continuous production due to improved technology is affecting healthcare and particularly genomics directly in terms of handling this ever-growing and an enormous amount of data. Estimates predict that genomics research will generate between 0.002 and 0.040 Zettabytes of data within the next decade, meaning requiring enormous amount of storage space only for this type of .[3]
In the evolution of genomic testing, the current Next-Generation Sequencing (NGS) methodologies have substantially increased the sequencing yield and produced a significantly more enormous amount of genomic data due to the lowered cost per base.[4] The switch from the previous Sanger sequencing, which allowed an analysis of few DNA fragments at a time (length < 1000bp), to the parallel sequencing of multiple genes (Next-generation sequencing, GS), in the early 2010s, is the reason that genomic laboratories now are handling significantly bigger genomic datasets. As the previous gold standard, Sanger sequencing could only produce a couple of Megabytes within a couple of months; today, these genetic laboratories face a data flow of multiple Gigabytes each week. The size of an Whole-Exome Sequencing (WES) dataset nowadays depends on the format, on particular attributes, such as the coverage of the genetic area, the sequencing kit, the sequencing machine, and the number of samples within the sequencing run.
All these developments put genomics and its laboratories under pressure to store these sensitive data, which needs to be held on average for at least 5 years by law, cost-efficiently to still run a healthy business.
Here is where one of the main benefits of the ISO/IEC 23092 MPEG-G standard comes into play: the efficient compression for genomic datasets. Thus far, the widely used formats such as FASTQ, BAM, or SAM, to mention a few, have been developed mainly in academic/research environments, and without following a solid standardization process, as typical for any other ISO standard in different industries.
The MPEG-G standard is built to compress the genomic data efficiently to provide the genomic laboratories with a format that allows them to store their data with fewer costs, increasing their business efficiency.
How does MPEG-G compress genomic data more efficiently?
The MPEG-G high compression efficiency is achieved using a hierarchical approach: at a first stage, the genomic information is split into so-called “descriptors”, that are representations of the different fields composing the genomic information (such as sequences, read names, quality values, …); descriptors are thus composed of symbols sharing a common probabilistic model and are specifically designed to maximize the efficiency of entropy coding (‘entropy’ in information theory is a variable that measures the average level of information in a signal). At the second stage, specific entropy coders, which have been finely optimized and tuned for maximum performances, are applied to descriptors to remove redundancies in a lossless way.
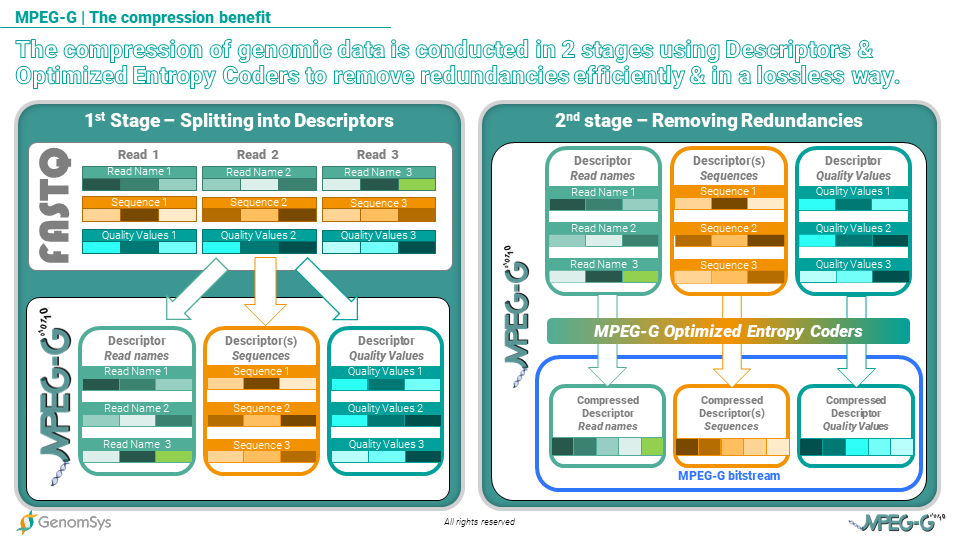
Graphic 1 – MPEG-G compresses genomic data in 2 stages more efficiently by using descriptors and optimized entropy encoders to remove redundancies losslessly.
For instance, the descriptor representing quality values is organized in a way that emphasizes the redundancy between the different symbols (each one representing a quality score as assigned to each base call by the sequencing machine) so that entropy coders can then easily remove such redundancy and thus achieve a high compression ratio.
What does that mean in practice for my laboratory?
Over the past year, Whole-Exome Sequencing has experienced a boost. The American College of Medical Genetics and Genomics(ACMG) recommended a Whole-Exome Sequencing (WES) or Whole-Genome Sequencing (WGS) as a test for specific inherited diseases because of the findings for a higher diagnostic yield.[5] As well did Germany in 2021, one of the major countries in Europe for genomic testing, reduce barriers in the reimbursement process for more extensive sized panels with more than 25kilobases including WES that now can be more easily prescribed without additional proposals to the health insurance providers.[6]
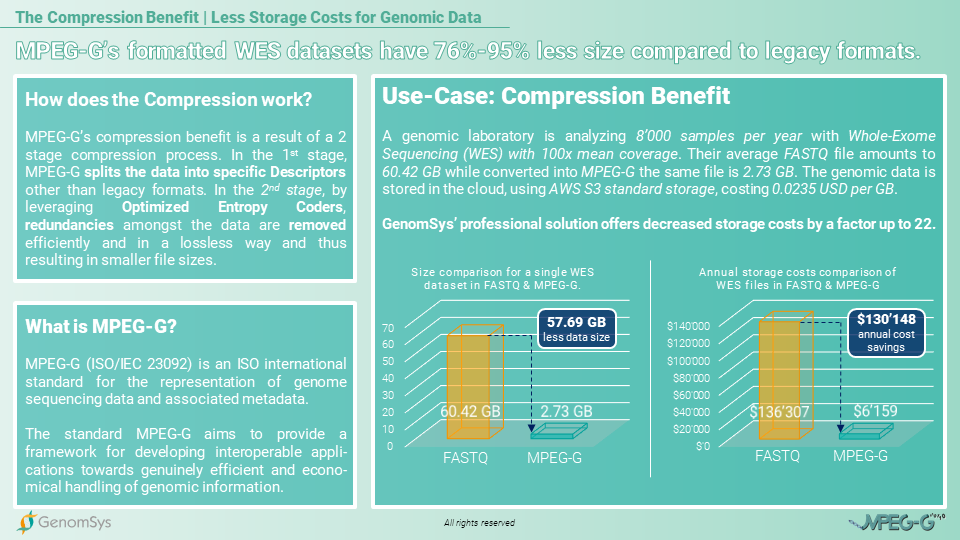
So for the following use case, we would like to showcase the compression benefit from MPEG-G for a medium-sized laboratory in Europe. This laboratory runs yearly 18’000 genetic samples in total, of which roughly 8’000 are examined by Whole-Exome Sequencing. The annual amount of data generated with WES, each dataset with a mean coverage of 100x, in the legacy format gzipped FASTQ will amount to 91.84 Terabytes of data. The exact amount of WES data in the MPEG-G format would generate 21.84 Terabytes of genomic data and thus deliver a compression rate more than 4 times higher than in the legacy format to the laboratory.[7]
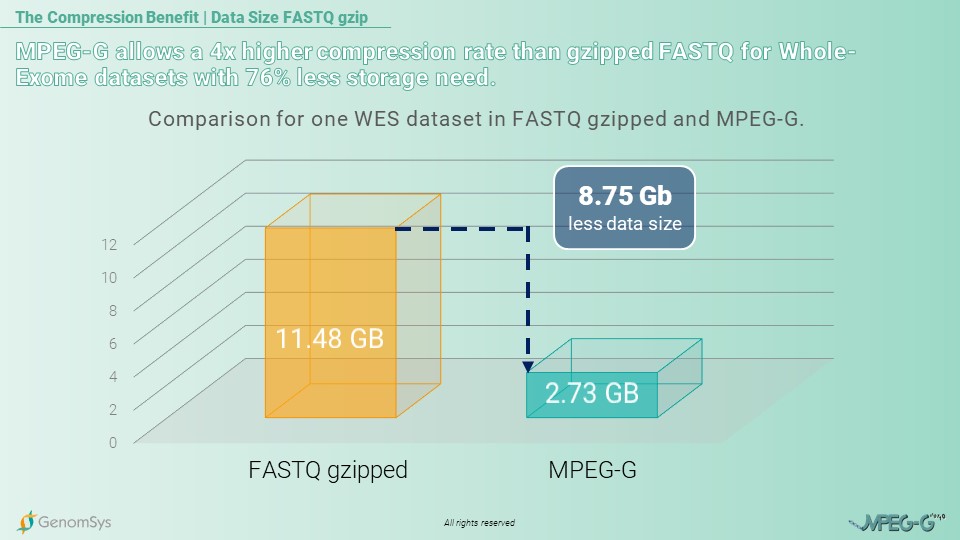
Graphic 2 – Comparison for the annual amount of data for WES genomic datasets in the gzipped FASTQ and MPEG-G format.[6]
If the same laboratory stores their 8’000 WES datasets in the FASTQ format, without compressing it through gzip, the compression benefit from MPEG-G would even increase significantly. The annual amount of data for FASTQ datasets would then amount to 483.36 Terabytes of data while MPEG-G would increase to 21.84 Terabytes per year. Still, delivering a compression rate more than 22 times higher compared to the legacy format FASTQ. [6]
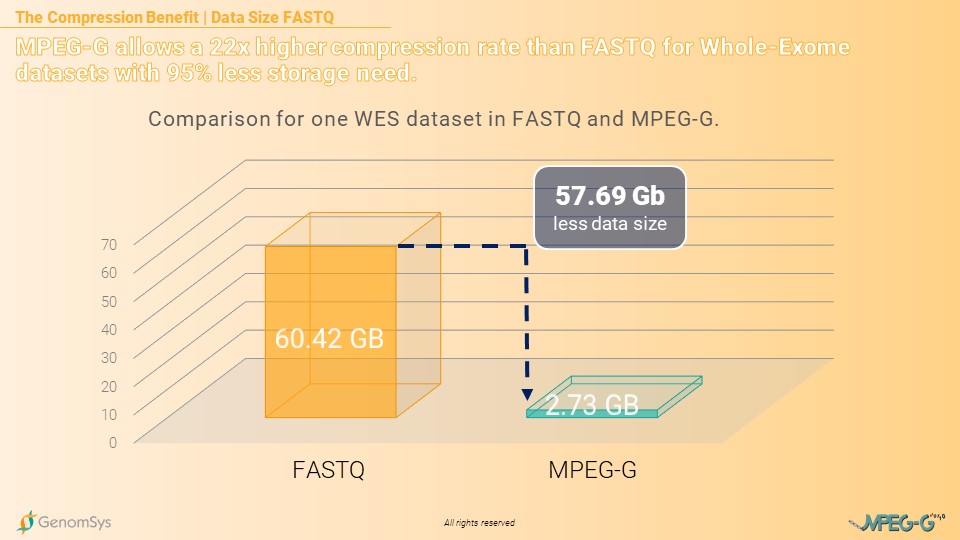
Graphic 3 – Comparison for the annual amount of data for WES genomic datasets in the FASTQ and MPEG-G format.[6]
The volume of genomic data is soaring, pushing traditional storage media to the limits of their capacity. Laboratories no longer have the space to store the mass of data locally. The solution is the cloud. And it has become more and more important amongst most industries as data storage is being outsourced to cloud service providers (e.g., Amazon or Microsoft) in the effort to save costs.[8]
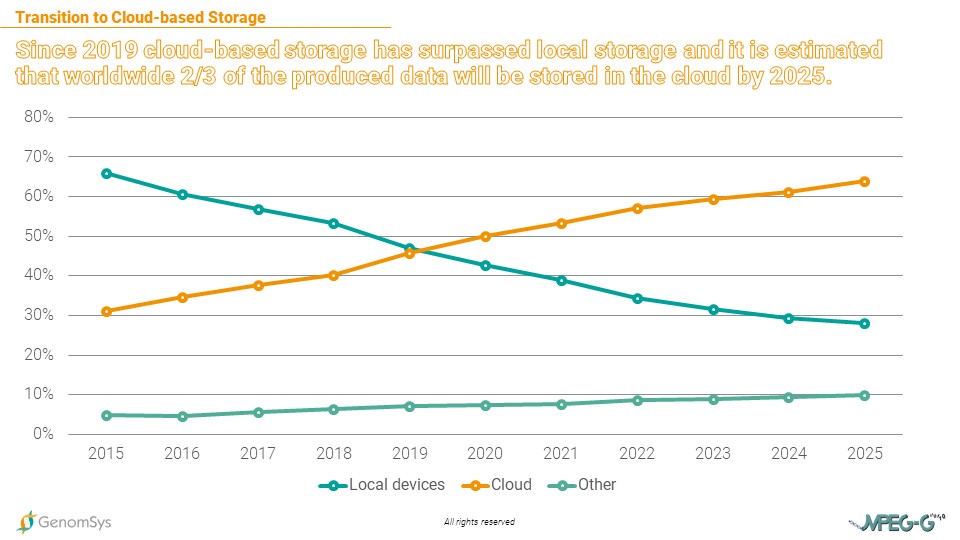
Graphic 4 – Shares of storage solutions in the storage of global data volume (in %).[7]
The cloud based storage is a trend that is important when talking about a reliable comparison for storage costs of genomic data. Considering that genetic data needs to be stored for at least 5 years on average by law in Europe, the storage costs for FASTQ gzipped formatted data, using a cloud service provider, will annually cost USD325’899 and for the whole 5 years USD129’495. In comparison, the MPEG-G formatted datasets would create annual costs of USD6’159 and for the entire legally prescribed period of 5 years USD30’795. Providing the laboratory cost savings of USD97’700 for storing 5 years the amount of WES datasets generated in 1 year.[9]
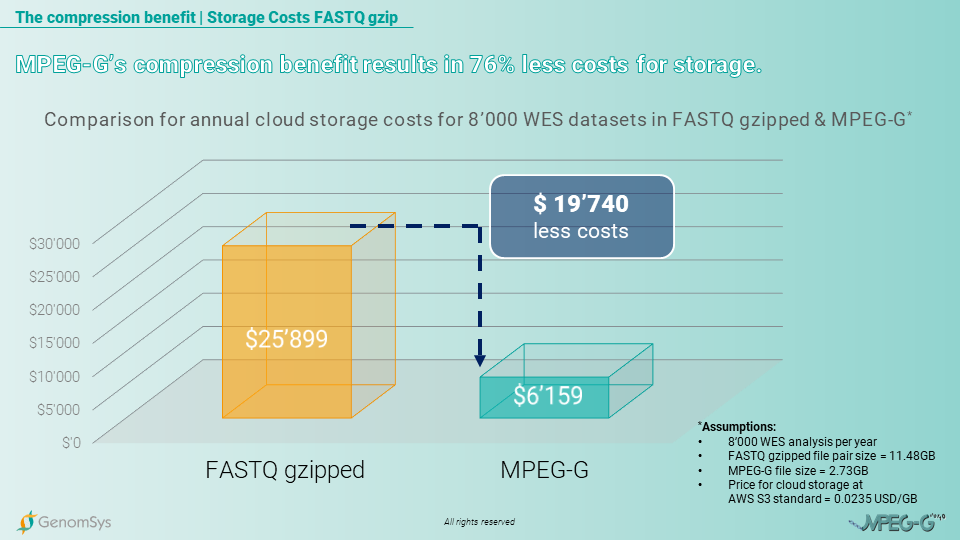
Graphic 5 – Comparison of annual cloud-based storage costs on AWS for WES genomic datasets in the FASTQ gzipped and MPEG-G format.[6] [8]
The storage benefit in costs even increases when comparing MPEG-G with FASTQ without being gzipped. The storage costs for the FASTQ formatted data, using a cloud service provider, will annually cost USD136’307 and for the whole 5 years USD681’535. In comparison, the MPEG-G formatted datasets would create annual costs of USD6’159 and for the entire legally prescribed period of 5 years USD30’795. The laboratory could save USD650’740 in storage costs by using MPEG-G for the 5 year period storing the annual amount of WES datasets generated.[8]
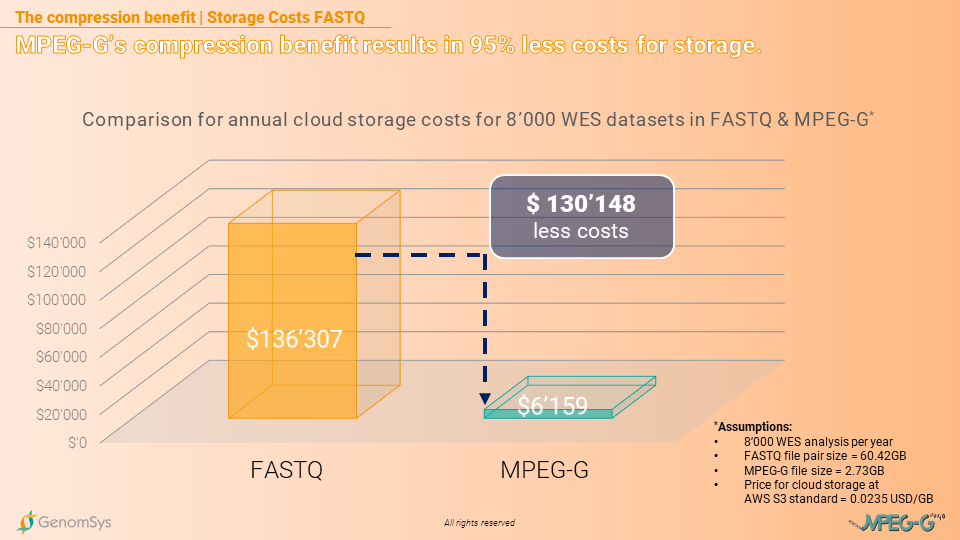
Graphic 6– Comparison of annual cloud-based storage costs on AWS for WES genomic datasets in the FASTQ and MPEG-G format.[6] [8]
GenomSys professional’s solution leveraging the compression benefit from MPEG-G
The CE marked GenomSys Variant Analyzer is natively running on the MPEG-G standard and allows converting seamlessly FASTQ data, coming directly from the sequencing machine, into MPEG-G before each analysis. This feature enables genomic laboratories to leverage the compression benefit from the unique ISO standard firsthand and save enormous storage costs. GenomSys helps organizations handle large amounts of genomic data to store them cost-efficiently and be equipped for the new era of genomic analysis with an ever-increasing volume of data.
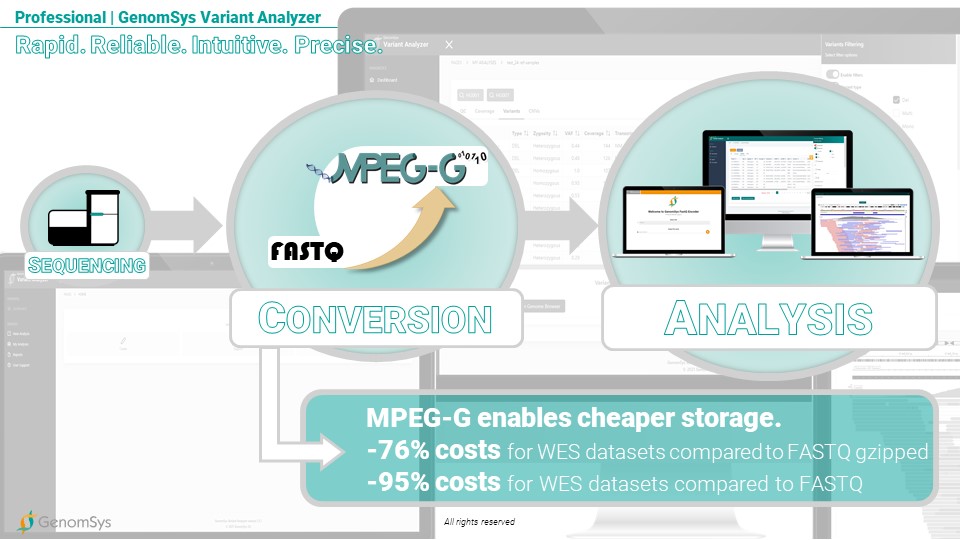
Graphic 7 – GenomSys professional solution workflow for genomic laboratories in their analysis of genomic data.
Also, working natively on MPEG-G, the entire analysis pipeline, including steps working on aligned data, can take advantage of all its features, from compression to Selective Access, thus minimizing the overall processing time and the storage and memory footprints. But don’t worry, we will cover the benefit of selective access in the next episode!
Please find our Fact-Card on MPEG-G with information on the Compression Benefit and a specific use-case in the following.
Stay tuned for our next article in the series “MPEG-G, can I eat it?” on the benefits of the Selective Access from the MPEG-G ISO standard next week.
By Lucas Laner and Daniele Renzi on March 9th, 2022.
References:
[1] Michael Kroker; Big Data sorgt schon 2016 für Speicher-Engpass; 2020 fehlt Speicher-Volumen von 6 Zetabytes (2015). https://blog.wiwo.de/look-at-it/2015/05/05/big-data-sorgt-schon-2016-fur-speicher-engpass-2020-fehlt-speicher-volumen-von-6-zetabytes/
[2] Christian Rusche; Datenmenge explodiert (2019). https://www.iwd.de/artikel/datenmenge-explodiert-431851/#:~:text=Die%20weltweite%20Datenmenge%20wird%20von,circa%20500%20Megabyte%20an%20Speicherplatz.
[3] Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, et al. (2015) Big Data: Astronomical or Genomical? PLoS Biol 13(7): e1002195. https://doi.org/10.1371/journal.pbio.1002195
[4] Buermans HPJ, den Dunnen JT. Next-generation sequencing technology: Advances and applications. Biochimica et Biophysica Acta (BBA) – Molecular Basis of Disease; From genome to function 2014 10/01;1842(10):1932-1941.
[5] Manickam, K., McClain, M.R., Demmer, L.A. et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet Med (2021). https://doi.org/10.1038/s41436-021-01242-6
[6] Institut des Bewertungsausschusses (2020): Entscheidungserhebliche Gründe zum Beschluss des Bewertungsausschusses nach § 87 Abs. 1 Satz 1 SGB V in seiner 547. Sitzung (schriftliche Beschlussfassung). Teil A. Zur Änderung des Einheitlichen Bewertungsmaßstabes (EBM mit Wirkung zum 1. Januar 2021. Verfügbar unter: https://institut-ba.de/ba/babeschluesse/2020-12-18_ba547_eeg_1.pdf
[7] In-House measurements (2022) of the same WES dataset with 100x coverage in each format: FASTQ gzipped file pairs= 11.48GB; FASTQ file pairs= 60.42GB; MPEG-G file=2.73G.
[8] Matthias Janson; 2020 überholt die Cloud lokale Speichermedien (2019). https://de.statista.com/infografik/18231/cloud-vs-lokaler-speicher/
[9] Amazon; Amazon S3 pricing (2022). https://aws.amazon.com/s3/pricing/?nc1=h_ls. Region of the server is Frankfurt price for S3 standard storage USD0.0235 per GB
Picture Source: Free-Photos/ pixabay