The healthcare system relies heavily on digital information systems to manage sensitive and interconnected medical information. However, the current IT framework lacks consistency, with the introduction of electronic health records (EHR) being inconsistent. This hinders the consolidation of medical data and metadata, such as audit trails of diagnostics, into one place for patients and professionals to follow easily. A data standard for genomic diagnostics could solve this issue, providing a transparent and traceable data trail.
The healthcare industry faces significant challenges due to technological advancements, increasing life expectancy, and a multitude of regulations and protocols, all while trying to maintain quality services with shrinking budgets.[1] Regulations and protocols often require extensive documentation, resulting in a cumbersome amount of manually filled-out papers and time-consuming information trails, particularly in a medical laboratory setting.
It’s common for people to experience at least one diagnostic error, including mistakes in lab tests conducted in medical laboratories. Even a tiny error can have negative consequences, such as misidentifying a patient or mixing up a specimen.[2] However, since mistakes are inevitable, having a well-functioning Quality Management System (QMS) in place is mandatory for medical laboratories. This system helps track laboratory processes, increasing the likelihood of improving the laboratory’s operations and minimizing the risk of new errors.
In this context, we aim to shed light on the laboratory workflow in genomics, the information flow along this path, and how the MPEG-G standard can serve as an intuitive container for all necessary information related to the trail of quality work in the laboratory.
What does the workflow look like in a genomics laboratory?
The workflow in a genomics laboratory involves multiple steps that transform the information in blood/saliva samples (containing our DNA) into actionable information for potential treatment decisions. This workflow is accompanied by a steady flow of information needed to deliver reliable results and ensure the highest quality level. With the increasing adoption of genetic testing in routine diagnostics, the entire process has become highly automated, including advanced technical support such as pipetting robots.
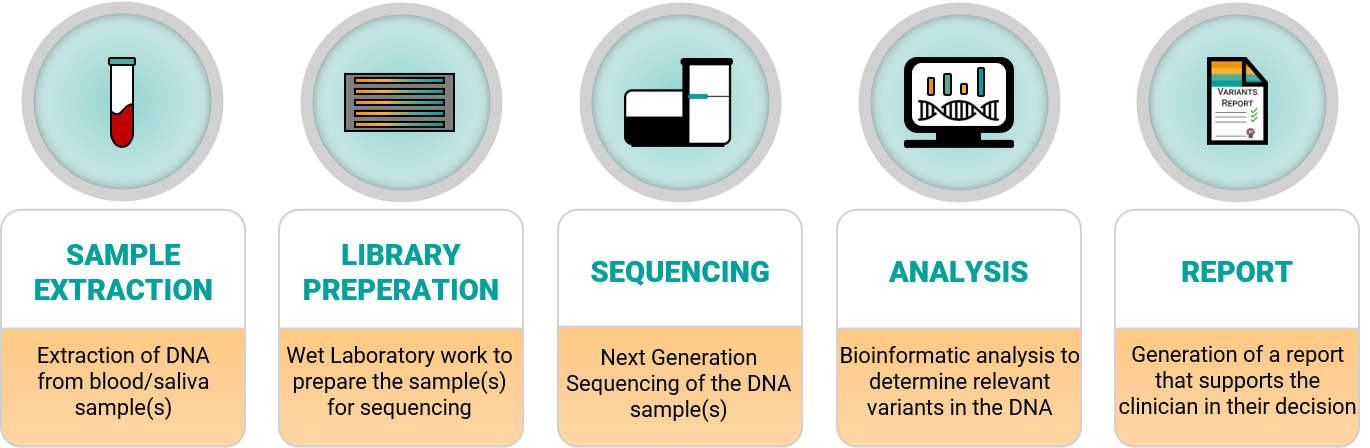
Figure 1 – Laboratory workflow including Sample Extraction, Library preparation, Sequencing, Analysis and Report generation.
What information is produced, and what IT systems are currently utilized throughout this process chain?
As our understanding of how our genome can contribute to specific diseases increased, the flow of information has expanded over time, particularly with the transition from Sanger Sequencing to Next Generation Sequencing. This evolution allowed for the sequencing of more extensive regions in our DNA and examining multiple samples in parallel, resulting in exponential growth in genomic data. While the complexity and need for automation in wet lab workflows are well-handled, interfaces among the information flow remain a delicate issue.
One of the significant issues is the heterogeneity of multiple software used in the genetic testing workflow. Most of them operate in their format regarding Laboratory Information Systems, making it challenging to implement a system combining Sequencing and Analysis data output. These outputs include up to five legacy formats (FASTQ, SAM, BAM, CRAM, and VCF) and must be integrated into a consistent stream of data, which is a time-consuming and challenging task.
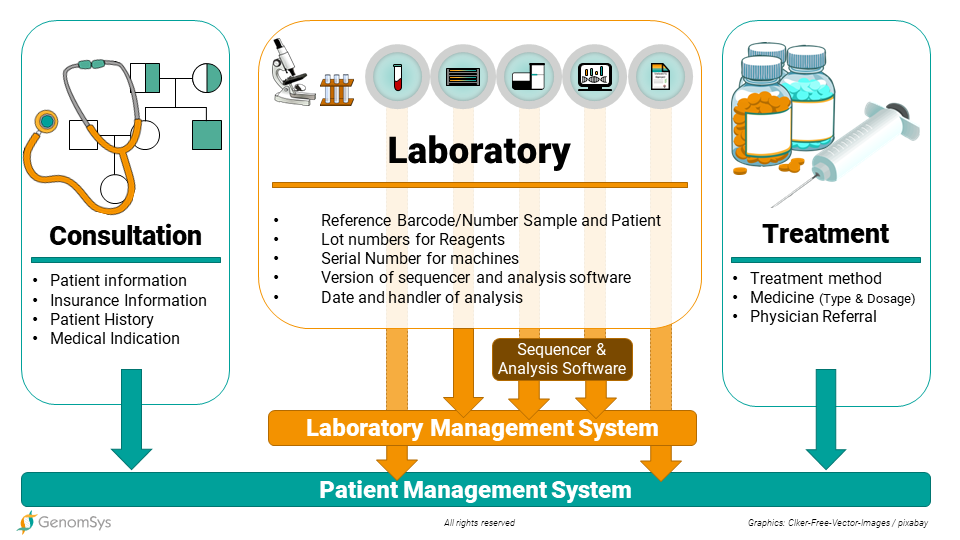
Figure 2 – Data flow in a genomic Laboratory including Patient Management System, Laboratory Information System and Analysis Software.
In what ways can the MPEG-G standard for genomic data assist laboratories in tracing their data trail?
The process of generating genomic data is a complex and sensitive undertaking that has led to an exponential increase in medical data in recent years. While bioinformaticians have been successful in managing this data and connecting raw sequencing data with variant annotations, integrating this data with other medical information has proven to be challenging due to the lack of standardization and interoperability in the currently used formats.
To address this issue, the Moving Picture Experts Group (MPEG) developed the MPEG-G standard (ISO/IEC 23092) for the representation of genome sequencing data and associated metadata. This standard ensures high interoperability and standardization, making it easier to connect genomic data with other medical information.
In addition to efficiently storing and handling genomic data, MPEG-G includes laboratory metadata such as patient history, reagents, sequencers, and analysis software, all relevant information in the laboratory workflow. The MPEG-G file for an individual patient becomes a single consistent file that is easy to retrieve and includes essential information on the data trail, including time stamp and responsibility tracking. This makes it easier to automate the audit trail and store it within a single file, simplifying the look-up process in case it is needed.
Video 1 – MPEG-G operates, as a container of genome sequencing data and associated metadata. The single file for a patient then simplifies keeping track of the data trail for Quality assurance and potential other QMS purposes.
By Lucas Laner on March 28th, 2023.
References:
[1] Prof Trish Greenhalgh and Dr Chrysanthi Papoutsi; Understanding Complexity in Health Systems: International Perspectives (unknown). https://www.biomedcentral.com/collections/complexity
[2] Robert Fenton; The 12 essentials of quality management in laboratory environments (2022). https://www.qualio.com/blog/quality-management-in-laboratory-environments
Picture Source: pexels / pixabay




