All 30 trillion cells in our body contain DNA be it bone cells, muscle cells, nerve cells or blood cells.[1] These cells had their building instruction stored in our genome and thus being, in general, all the same, as 99.9% of our genetic makeup is identical, the 0.1% delivers differences that make mass medicine complex.[2]
Let’s take blood cells, for example. Today, there are 43 blood systems recognized by the International Society of Blood Transfusion (ISBT). All of these help classify the blood that is running through our bodies.[3] Most of them categorize the differences in 345 known antigens of our red blood cells (RBC) and are genetically determined by 48 known genes.[4] The most commonly known are the AB0 system and those of the rhesus factors.
The characterization of the blood is needed in the case of blood transfusion to ensure the compatibility between the donor blood and the recipient. If this were not the case, the antibodies would “attack” the transfused erythrocytes and clump together. This could have life-threatening consequences. So before operations, the blood groups are always tested, which takes about 45 minutes in the laboratory. If someone needs a blood unit more quickly, blood group 0 negative is always administered due to the compatibility with the other groups.[5]
In the following, we would like to highlight the three most important blood type classification systems: the AB0-, the Rhesus-, and the Kell systems and give an outlook on how genomic testing might be a more efficient approach in the future to analyze the characteristics of our blood in the future.
The AB0 system
In 1900, Karl Landsteiner discovered and developed the first human blood group system and called it the AB0-system, which is, to this day, the most important blood system when talking about blood transfusion. This system classifies the different membrane receptors of the erythrocytes – the scientific name for red blood cells – with different letters.[6]
Globally, blood types A and 0 are the most common, but the distribution of the four blood types varies from region to region. For example, B is most common in most areas of Asia, A in Europe, and blood type 0 in South America and some African countries.[7]
| A | B | 0 | AB | |
| Worldwide[8] | 43% | 11% | 41% | 5% |
| Switzerland[9] | 45% | 9% | 41% | 5% |
| Italy[9] | 42% | 9% | 46% | 3% |
| Spain[9] | 43% | 10% | 44% | 3% |
| Germany[9] | 43% | 11% | 41% | 5% |
Figure 1 – Blood type distribution worldwide, Switzerland, Italy, Spain and Germany for the Blood group system AB0.
But how did the human species develop these different membrane receptors that Landsteiner discovered in his experiments in the late 19th century and for which he won the Nobel prize in medicine?6]
Polymorphisms within the AB0 gene determine the different types of the AB0 blood group system. Evolutionarily, given that blood types A (rs8176746) and B (rs8176747) share polymorphisms with other primates, it seems that those two might be the oldest blood types within the AB0 blood group system. This assumption is based on the fact that blood type 0 is caused more commonly – but not exclusively – by a single base deletion (rs8176719) in the genotype.[10] And despite the possibility that an insertion created the blood types A and B after the blood type 0, it seems far more likely – due to the greater frequency for deletion compared to insertion within the DNA – that a later deletion led to the formation of blood type 0.[11]
The blood group inheritance of the AB0 system follows the Mendelian inheritance rules, which are based on the findings of the natural scientist Gregor Mendel. The prerequisite for this is that a gene determines the inheritance of the blood group. Every person has two copies of this gene (alleles), one inherited from the father and the other from the mother, and the two copies might differ in terms of sequence due to the presence of variants. The combination of these two alleles determines a person’s blood group genotype.[12]
In terms of the AB0 blood group system, each person has two alleles of the three traits A, B, and 0. Accordingly, AA, BB, 00, A0, B0, and AB are possible genotypes. In inheritance patterns, alleles A and B are equivalent to each other, while they are dominant to allele 0. This means, for example, that blood type A results from genotype A0 and blood type AB from genotype AB. The blood type 0 results only in the case of a genotype 00, i.e., if both alleles inherited from the parents have the same so-called recessive trait 0.[12]
| A | B | 0 | AB | |||
| Genotypes | A0 | AA | B0 | BB | 00 | AB |
Figure 2 – The potential genotypes for the four different blood types in the AB0 blood system.[12]
How does the AB0 Blood groups test work?
The ABO blood type test is widely used in hospitals as a bedside test for rapid blood group verification. It is mandatory before the transfusion of RBC to avoid transfusion errors.[13]
The ABO characterization test uses a simple card with two or sometimes three test fields (the third is for the rhesus factor and will be explained in detail later on). Each test field contains a serum with either anti-A antibodies or anti-B antibodies. One drop of blood is added to each field. The antibodies’ serum and the antigens’ blood drops are then mixed. And based on agglutination (clumping) or non-agglutination, due to the antigen-antibody reaction, the respective blood type is determined.[13]
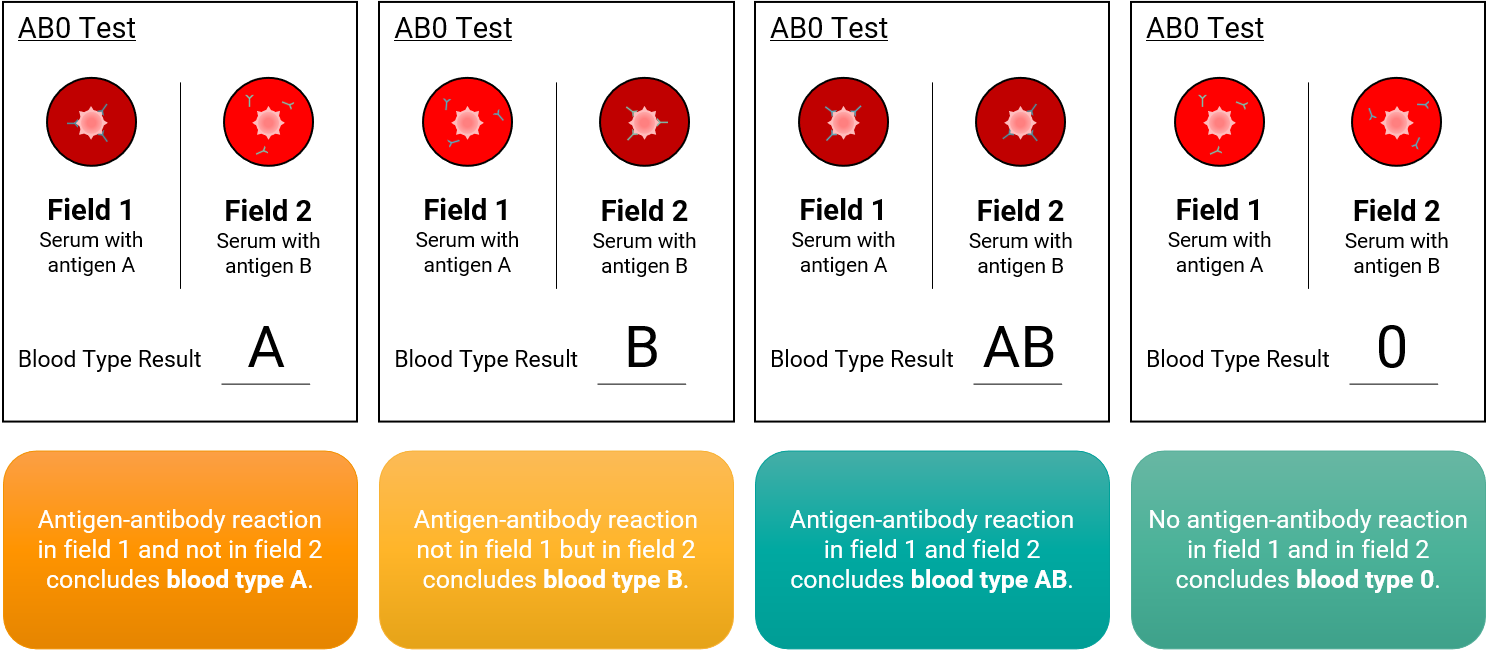
Figure 3 – Mechanism and application of the AB0 blood group card test. Field 1 contains serum with anti-A antibodies, “clumping” of blood determines Blood type A or AB. Field 2 contains anti-B antibodies; if agglutination occurs, the blood type is either B or AB. No “clumping” suggests the blood type is 0.[13]
The rhesus system
The rhesus (Rh) blood group system is one of the most complex blood groups known in humans, discovered more than 80 years ago by – again – Landsteiner and his students Levine and Wiener. In his experiments, Landsteiner wanted to produce antibodies against erythrocytes of the rhesus monkey in rabbits and guinea pigs. In the process, he discovered that the same antibodies also agglutinate erythrocytes in humans.[14]
The Rh system distinguishes between Rh positive (Rh+) and Rh negative (Rh-). Rh+ means that a particular antigen is present on the surface of the blood cells; persons who do not possess this antigen are, therefore, RH-. The rhesus system includes a group of related proteins, the five most essential representatives (C, c, D, E, e), called rhesus factors.[14] The most important rhesus factor has the abbreviation “D”.[15] Today, it is still the second most important in transfusion medicine, only trailing the AB0 blood group system. The complexity of the Rh blood group antigens begins with the closely linked highly polymorphic genes (RHD and RHCE) that encode them. Numerous genetic rearrangements between them have produced hybrid Rh genes that encode a myriad – 56 to date – of distinct Rh antigens.[16]
The significance of the Rh blood group is related to the fact that the Rh antigens are highly immunogenic, causing an immune response. In the case of the D antigen, individuals who do not produce the D antigen will produce anti-D if they encounter the D antigen on transfused RBCs (causing a hemolytic transfusion reaction, HTR) or on fetal RBCs (causing HDN). For this reason, the Rh status is routinely determined in blood donors, transfusion recipients, and mothers-to-be.[15]
The rhesus factor is inherited according to Mendelian rules. The mode of inheritance is dominant-recessive, i.e., the expression of the factor is dominant to the rhesus-negative phenotype. The majority of Central Europeans are Rh-positive. The proportion of rhesus-negative people in the population of Central Europe is between 15 and 17 %. On other continents, this proportion is significantly lower. The indigenous peoples of America, Australia and East Asia, for example, are 100 % rhesus-positive.[15]
How does the rhesus system test work?
Usually, the rhesus classification is done together with the ABO blood type through a simple test card as a bedside test for rapid blood group verification. In this case – as previously mentioned – the third test field is used to test for the rhesus factor.
The rhesus test field contains a serum with anti-D antibodies. One drop of blood is added to each field. The antibodies’ serum and the antigens’ blood drops are then mixed. And based on agglutination or non-agglutination, the respective rhesus factor( Rh+ or Rh-) is determined due to the antigen-antibody reaction.
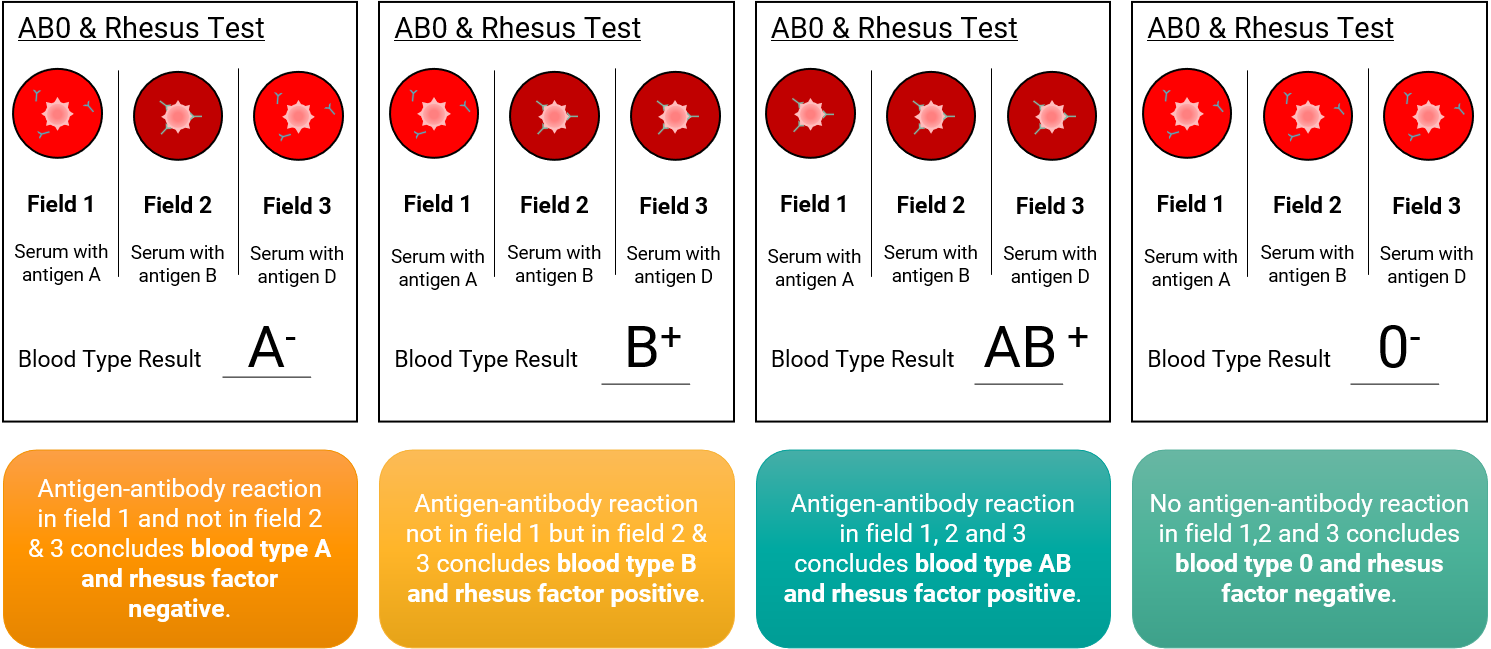
Figure 4 – Mechanism and application of the Combination blood group card test for AB0 blood group and Rhesus system. Field 1 contains serum with anti-A antibodies, “clumping” of blood determines Blood type A or AB. Field 2 contains anti-B antibodies; if agglutination occurs, the blood type is either B or AB. Field 3 contains anti-D antibodies; if agglutination occurs, the rhesus factor is present, meaning rhesus-factor negative. No “clumping” for fields 1 and 2 suggests the blood type is 0 and for field 3 rhesus factor negative.
The Kell system
After the previously described and best-known AB0 and Rhesus systems, the Kell system, also called the Kell-Cellano system, is the third most relevant blood group system test in routine diagnostics.
In 1946, the attending physician Sir Robin Coombs discovered an incompatibility between the blood groups of his patient and her daughter, that was born with Morbus haemolyticus neonatorum. The new antibody and the corresponding antigen were given the patient’s name: Mrs. Kelleher. A later publication by Ph. Levine et al. found the antibody postulated for the corresponding allele in the serum of another patient, Mrs. Nocella, who had given birth to a child with mild hemolytic neonatorum disease. According to the mother’s name, the corresponding antigen should have been called Nocella. However, due to a probable letters switch, Nocella turned into Cellano. Accordingly, the classifying system is now called the Kell-Cellano system.[17]
The Kell system distinguishes between 37 antigens. This system’s most important antigens are K (KEL1, Kell) and k (KEL2, Cellano). The responsible known genes for the antigen Kell are KEL, ECE3 and CD238 as well are regions of the gene KEL responsible for the antigen Cellano.[18] Since the gene KEL plays a role for both antigens, KEL1 and KEL2 are called antithetic antigens since they can represent each other in the case of polymorphisms of the structural gene KEL.[19]
The worldwide blood group distribution is quite one-sided: 92% of people are Kell-negative. As donation recipients, these people may only receive Kell-negative blood for a safe blood transfusion. Kell-positive patients, on the other hand, can receive both Kell-positive and Kell-negative blood preparations.[20]
Especially in the context of pregnancy, the Kell system plays an important role: because the effect of a Kell incompatibility between the mother (Kell factor negative with antibodies) and child (Kell positive) can, in severe cases, lead to diseases of the child such as anemia in the yet unborn child. Therefore, the compatibility of the most critical blood group characteristics in a blood transfusion is particularly important for young women.[20]
The characteristics of our blood lie in our genes; why don’t we analyze DNA to receive the information then?
As mentioned before, the building information for all cells, including blood cells, lies within our DNA. Variations within the code of life determine each of us individually. For some genes, as shown for the significant blood group systems previously, this information can be extracted via a DNA analysis. This raises the question of why not simply use genetic testing to characterize all 43 blood groups at once, which today is not yet used for the characterization of the blood.
In the past, genetic testing, especially DNA sequencing, was very costly. Compared to the long-time established serological tests, due to their wide applications and simplistic setup, genetic testing for the characterization of the blood was not efficient from an economic standpoint. But technological breakthroughs over the past decade drastically improved genetic testing methods, supporting the automation level of sequencing processes and reducing the cost per analysis. Next-generation sequencing (NGS)-based methods, introduced in 2005, allow for parallel sequencing of multiple genes, such as the whole protein-coding region (whole-exome sequencing, WES) or whole genomes (whole-genome sequencing, WGS).[21]
Nowadays, the actual sequencing costs less and more individuals already have their DNA sequenced or will have it sequenced if the current projections from different initiatives become a reality, e.g., the National Human Genome Research Institute envisions that everyone in the US will have their genome sequenced.[22] The benefits – receiving the entire overview of the blood characteristics – will then outweigh the costs.
And this delivers one of the apparent advantages of analyzing our DNA to characterize the blood. Instead of running multiple individual tests – up to 43 at max – single genetic testing could deliver the full characteristics of the blood.
| Blood group system | Involved Gene(s) | No. of Antigens |
| ABO | ABO | 4 |
| Rhesus | RHD, RHCE | 56 |
| Kell-Cellano | KEL | 36 |
| MNS | GYPA, GYPB, (GYPE) | 50 |
| P1PK | A4GALT | 3 |
| Lutheran | BCAM | 27 |
| Lewis | FUT3 | 6 |
| Duffy | ACKR1 | 5 |
| Kidd | SLC14A1 | 3 |
| Diego | SLC4A1 | 23 |
| Yt | ACHE | 5 |
| Xg | XG, CD99 | 2 |
| Scianna | ERMAP | 9 |
| Dombrock | ART4 | 10 |
| Colton | AQP1 | 4 |
| Landsteiner-Wiener | ICAM4 | 3 |
| Chido/Rodgers | C4A, C4B | 9 |
| H | FUT1; FUT2 | 1 |
| Kx | XK | 1 |
| Gerbich | GYPC | 13 |
| Cromer | CD55 | 20 |
| Knops | CR1 | 12 |
| Indian | CD44 | 6 |
| Ok | BSG | 3 |
| Raph | CD151 | 1 |
| John Milton Hagen | SEMA7A | 8 |
| I | GCNT2 | 1 |
| Globoside | B3GALNT1 | 2 |
| Gill | AQP3 | 1 |
| Rh-associated glycoprotein | RHAG | 4 |
| FORS | GBGT1 | 1 |
| JR | ABCG2 | 1 |
| LAN | ABCB6 | 1 |
| Vel | SMIM1 | 1 |
| CD59 | CD59 | 1 |
| Augustine | SLC29A1 | 4 |
| Kanno | PRNP | 1 |
| SID | B4GALNT2 | 1 |
| CTL2 | SLC44A2 | 2 |
| PEL | ABCC4 | 1183 |
| MAM | EMP3 | 1 |
| EMM | PIGG | 1 |
| ABCC1 | ABCC1 | 1 |
Figure 5 – Overview of 43 blood group characterization systems and their corresponding genes.[23]
Although the duration of the analysis may be longer since the DNA has to be sequenced and analyzed, this analysis only has to be performed once. As mentioned before, if more and more individuals already have their genomes sequenced, without sequencing and solely analyzing their sequenced DNA could accelerate characterizing the blood.
Another reason there has not been any routine genetic testing for the characterization of the blood was the high complexity of the genetic composition of the regions on the DNA. Most regions on the DNA are so homologous, having the same or very similar sequence, complicating the analysis. This complexity – e.g., the most common AB0 system – is a clear barrier, as there is no margin for error in blood transfusion.[24]
However, hope is on the horizon that in the future, via genetic testing, a fast and efficient determination of our blood based on our genes will be possible. The continuous development of bioinformatic pipelines that provide reliable results for the more complex genotype relationships for the different blood group systems gives reason for that.
By Lucas Laner on December 13th, 2022.
References:
[1] Jacquelyn Cafasso; How Many Cells Are in the Human Body? Fast Facts (2018). https://www.healthline.com/health/number-of-cells-in-body
[2] National Human Genome Research Institute; Genetics vs. Genomics Fact Sheet (2018). https://www.genome.gov/about-genomics/fact-sheets/Genetics-vs-Genomics
[3] Mitra R, Mishra N, Rath GP. Blood groups systems. Indian J Anaesth. 2014 Sep;58(5):524-8. doi: 10.4103/0019-5049.144645. PMID: 25535412; PMCID: PMC4260296.
[4] Catherine Hyland, Christoph Gassner; Red Cell Immunogenetics and Blood Group Terminology (2021). https://www.isbtweb.org/isbt-working-parties/rcibgt.html
[5] Vera Denkhaus; Blutgruppen bestimmen: So einfach finden Sie Ihre heraus (2022). https://www.morgenpost.de/vermischtes/article235672813/blutgruppe-blutspende-drk-corona-herausfinden-test.html
[6] Karl Landsteiner Gesellschaft; Wissenswertes: Dr. Karl Landsteiner – Arzt und Bakteriologe (undefined). https://www.karl-landsteiner.at/wissenswertes.html
[7] Dennis O’Neil; Distribution of Blood Types (2012). https://www2.palomar.edu/anthro/vary/vary_3.html
[8] DRK-Blutspende Team; Blutgruppen-Verteilung in der Bevölkerung (2020). https://www.blutspende.de/magazin/von-a-bis-0/blutgruppen-verteilung-in-der-bevoelkerung
[9] World Population Review; Blood Type by Country 2022 (2022). https://worldpopulationreview.com/country-rankings/blood-type-by-country
[10] Iain Mathieson; Blood groups in ancient Europe (2017). https://mathii.github.io/2017/09/21/blood-groups-in-ancient-europe
[11] Fan Y, Wang W, Ma G, Liang L, Shi Q, Tao S. Patterns of insertion and deletion in Mammalian genomes. Curr Genomics. 2007 Sep;8(6):370-8. doi: 10.2174/138920207783406479. PMID: 19412437; PMCID: PMC2671719.
[12] Ilona Miko; Genetic Dominance: Genotype-Phenotype Relationships (2008). https://www.nature.com/scitable/topicpage/genetic-dominance-genotype-phenotype-relationships-489/
[13] Daniela Niehaus; ABO-Identitätstest (2014). https://flexikon.doccheck.com/de/ABO-Identit%C3%A4tstest
[14] Conatex-Didactic Lehrmittel GmbH; Blutgruppen-Bestimmung im ABO-System (undefined). https://www.conatex.com/media/experiments/VADE/VADE_Biologie_Blutgruppen.pdf
[15]Georg Graf von Westphalen, Pascal Henrich, Ralf Huch, Frank Antwerpes; Rhesusfaktor (2021). https://flexikon.doccheck.com/de/Rhesus-System
[16] Dean L. Blood Groups and Red Cell Antigens [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2005. Chapter 7, The Rh blood group. Available from: https://www.ncbi.nlm.nih.gov/books/NBK2269/
[17] Markus Böck ; Transfusionsmedizin Basiswissen für Studierende – Das Kell-System (undefined). https://www.transfusionsmedizin-vorlesung.de/vorlesung-transfusionsmedizin/blutgruppen-erythrozyten-iii/das-kell-system/
[18] R. R. A. Coombs, A. E. Mourant, R. R. Race: A new test for the detection of weak and incomplete Rh agglutinins. In: Br J Exp Pathol. 26, 1945
[19] K. Kleesiek, C. Götting, J. Diekmann, J. Dreier und M. Schmidt; Lexikon der Medizinischen Laboratoriumsdiagnostik: Antithetische Antigene (2018). https://www.springermedizin.de/emedpedia/lexikon-der-medizinischen-laboratoriumsdiagnostik/antithetische-antigene?epediaDoi=10.1007%2F978-3-662-49054-9_252
[20] DRK-Blutspendedienste; Kell-System in der Blutgruppenbestimmung (2020). https://www.blutspende.de/magazin/von-a-bis-0/kell-system-in-der-blutgruppenbestimmung
[21] Jamuar SS, Tan EC. Clinical application of next-generation sequencing for Mendelian diseases. Hum Genomics 2015 Jun 16;9:10-015-0031-5.
[22] National Human Genome Research Institute; Bold Predictions for Human Genomics by 2030: An NHGRI Seminar Series (2022). https://www.genome.gov/event-calendar/Bold-Predictions-for-Human-Genomics-by-2030
[23] International Society of Blood Transfusion; Table of blood group systems v10.0 (2021). https://www.isbtweb.org/asset/3BBBD515-1BF4-4EF4-80E9858F10C2B1AB/
[24] Daniels G. An overview of blood group genotyping. Ann Blood 2021.





