Over the past decade, genomics has progressed rapidly, and Next-Generation Sequencing (NGS) has been one of the main drivers for this circumstance. A growing pile of genomic data is continuously generated through the acceleration of sequencing DNA, from taking a couple of months for a couple of exons to having an entire genome sequenced within a couple of days.. Therefore, genomics now faces the same challenges as any other field handling Big Data, moreover complicated by the truly personal information comprised by our DNA [1].
The progress of genomics has been the result of continuous research processes that would have been impossible without sharing data and information amongst researchers and institutions. The COVID-19 pandemic was a prime example of how a worldwide effort in scientific research in finding an antidote against the coronavirus is possible and to a certain degree successful but needs a certain level of interoperability.
But interoperability is not a given in healthcare or genomics: data is out there, but it doesn’t mean it is accessible or usable. In fact, in genomics, you find a vast number of data stored in different setups, with data repositories containing valuable information often locked away in a sort of digital solitary confinement [2]. Although interoperability might not be available due to that circumstance, it is only possible if multiple, time-consuming steps to prepare the data for sharing are done. True interoperability looks different.
The current genomic testing relies on multiple legacy data formats, each with a different purpose. These differences result in several data files for each genomic dataset per individual. Besides the additional needed storage capabilities, this circumstance introduces an additional layer of complexity when sharing data. As the approach with Electronical Health Records (EHR) (see our article for more information on EHR and eHealth), one “file” containing all the medical history of a patient, a single unified syntax in an open format for genomic data representation could dissolve the complexity. The result is a single file containing all the genomic information of an individual, making conversion between data formats a thing of the past when exchanging data for medical advances, ultimately saving valuable time.
Additionally to the current status quo for the silos of genomic information, the fact that DNA information is highly sensitive, sharing genomic data requires a much higher level of data protection. These data privacy protection legislation, although very important, vary from country to country in Europe alone, and it is even more difficult on a more global scale to share genomic data. This patchwork of legislation leads to a slowdown in medical progress and constrains the full potential of research.
Technology can be the answer to building a convenient bridge between protecting genomic data privacy and offering a secure way to share information for research purposes. The needed Data security can be achieved by blocking unauthorized users with various techniques like access control, homomorphic encryption, and Secure Multiparty Computation. Besides this, there is still a need to anonymize the data, which can be achieved by suppressing certain SNVs and other randomizing mechanisms. In the future, opportunities could be created for controlling the data flow to increase privacy as well. The focus lies on enabling individuals to own, track and potentially even profit from their genomic data [3].
At GenomSys, we want to address these challenges head-on. We rightfully want to establish an ecosystem in genomics: truly interoperable and highly protective of the valuable data that is our DNA. Our solutions, GenomSys Variant Analyzer, software for professionals to analyze genetic data, and GenomSys Codec Suite, a tool to transform legacy genomic data formats into the more efficient and future-proof MPEG-G format, reflect that.
What does that mean in practice? Let’s have a look at a use case.
In the example below, GenomSys’ ecosystem is based on standardized and interoperable technology that enables clinicians to identify faster and more efficiently data that is needed to make a new genetic analysis. Data hidden in non-interoperable repositories will never be searchable or usable, thus not really actionable.
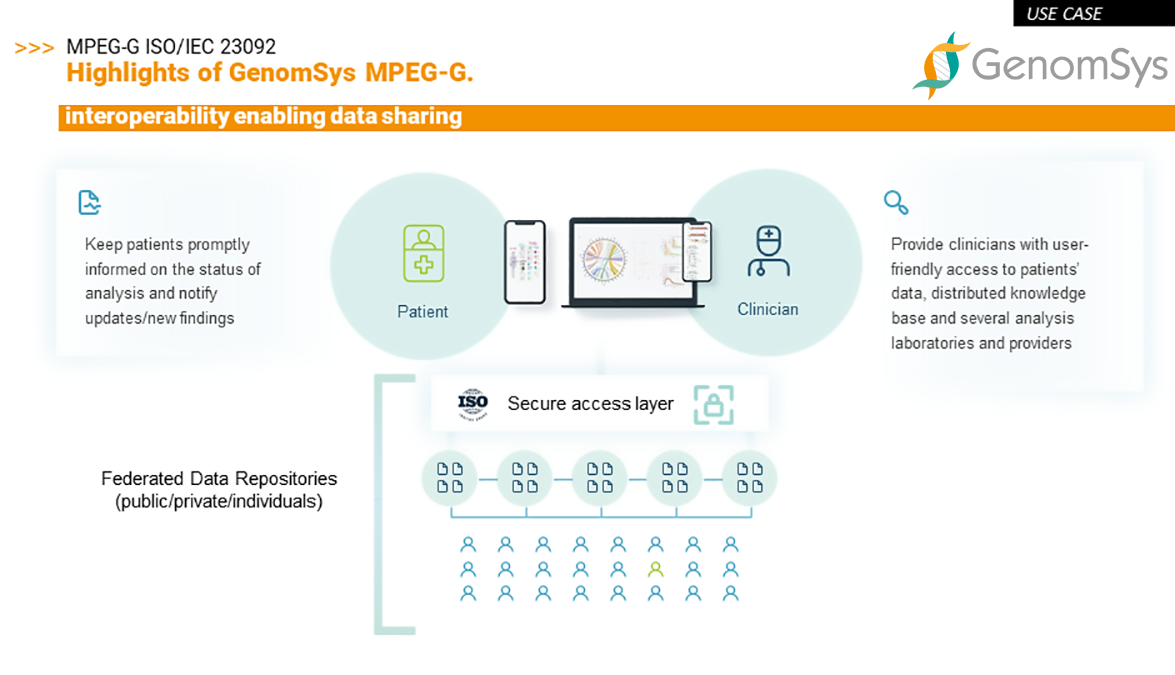 We believe that research and healthcare in the future – as we had the glimpse already in the COVID-19 pandemic – will be even more connected and reliable on a rapid interaction amongst researchers, including sharing genomic data. We want to support this future development of state-of-the-art tools for professionals to support them in their daily diagnostic routine and contribute – through the standard – to genomic research to ultimately leverage the human genome and its containing information to improve patients’ lives.
We believe that research and healthcare in the future – as we had the glimpse already in the COVID-19 pandemic – will be even more connected and reliable on a rapid interaction amongst researchers, including sharing genomic data. We want to support this future development of state-of-the-art tools for professionals to support them in their daily diagnostic routine and contribute – through the standard – to genomic research to ultimately leverage the human genome and its containing information to improve patients’ lives.
By Lucas Laner on December 21, 2021.
References:
[1] McGuire, A.L., Gabriel, S., Tishkoff, S.A. et al. The road ahead in genetics and genomics. Nat Rev Genet 21, 581–596 (2020). https://doi.org/10.1038/s41576-020-0272-6[2] Baha Zeidan. COVID-19 Highlights the Need for Interoperability in Healthcare
[3] Bonomi, L., Huang, Y. & Ohno-Machado, L. Privacy challenges and research opportunities for genomic data sharing. Nat Genet 52, 646–654 (2020). https://doi.org/10.1038/s41588-020-0651-0
Picture: Pete 





