Today in our series “MPEG-G, can I eat it?” we want to shine a light on how the MPEG-G standard in its nature, being developed by the International Organization for Standardization (ISO), is meant to be highly interoperable. We want to explain why interoperability for genomic data is needed and how it is provided by MPEG-G, with a practical example of how this interoperability benefit can directly affect the field of genomics.
If you missed our previous articles, you can find them here:
- MPEG-G, can I eat it? – Definition & History
- MPEG-G, can I eat it? – The Compression Benefit
- MPEG-G, can I eat it? – The Selective Access Benefit
Sharing is Caring – Interoperability is key for a steady medical progress
Genomics has progressed rapidly over the past decade, propelled by the pace of Next-Generation Sequencing (NGS) technology paired with Bioinformatics. A growing pile of genomic data is continuously generated through the acceleration of sequencing DNA, as today we can have an entire genome sequenced within a couple of days. Therefore, genomics now faces the same challenges as any other field handling Big Data, moreover complicated by the truly personal information comprised by our DNA.[1]
Genomics has evolved as a reliable and helpful resort in healthcare based on continuous research processes that would have been impossible without sharing data and information amongst researchers and institutions. The COVID-19 pandemic was a prime example of how a global effort in scientific research in finding an antidote against the coronavirus is possible and successful. But achieving that requires a level of interoperability, meaning the ability for various healthcare information technology to exchange, interpret and use data cohesively.
The mRNA vaccines from this intercontinental medical research have brought genomics and genomic data into the minds of the broad public. Bernasconi et al., showed how interoperability would be essential for the velocity of any future progress in healthcare. As leveraging corresponding health data ranging – particularly for COVID19 – viral genomics, clinical phenotypes, and human genomics genotypes could result in a reliable and helpful weapon in battling future genomic-related diseases.[2]
But high interoperability is not yet a given in healthcare or genomics: data is out there, but it doesn’t mean it is accessible or usable. In fact, in genomics, you find a vast number of data stored in different setups, with data repositories containing valuable information often locked away in a sort of digital solitary confinement.[3] Although interoperability might not be available due to that circumstance, it is only possible if multiple, time-consuming steps to prepare the data for sharing are done. True, efficient interoperability looks different.
The current genomic testing relies on multiple legacy data formats, each with a different purpose, leading to several genomic data files for each individual. Besides the additional needed storage capabilities – more information on MPEG-G’s storage benefit here – the variety of formats introduces an extra layer of complexity when sharing data.
Additionally to the current status quo for the silos of genomic information amongst all these legacy formats, the fact that DNA information is highly sensitive, sharing genomic data requires a much higher level of data protection. These data privacy protection legislation, although very important, vary from country to country in Europe alone, and it is even more difficult on a more global scale to share genomic data. More on the Security of genomic data we’ll explain next week.
What makes the MPEG-G format data more interoperable?
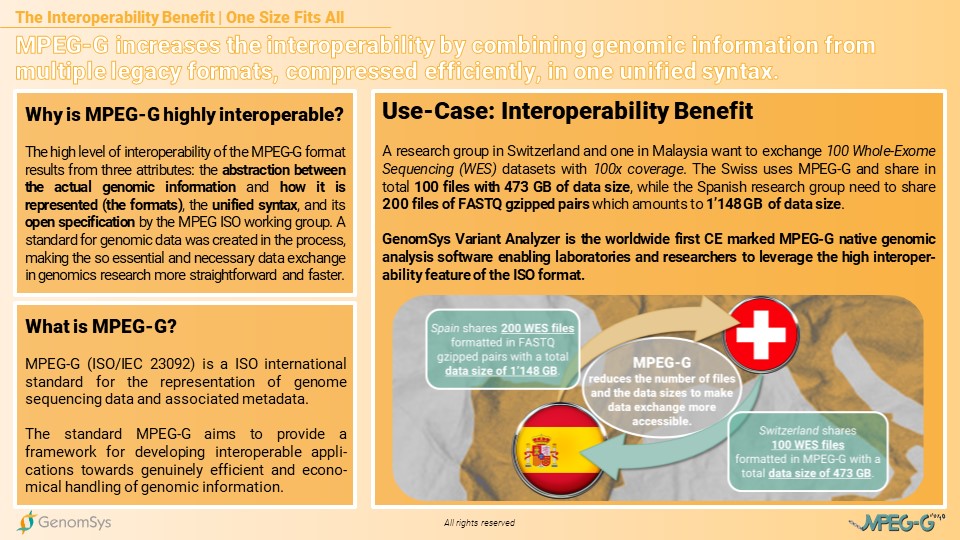
The high level of interoperability of the MPEG-G format results from three attributes: the abstraction between the actual genomic information and how it is represented (the formats), the unified syntax, and its open specification by the MPEG ISO working group. A standard for genomic data was created in the process, making the so essential and necessary data exchange in genomics research more straightforward and faster.
MPEG-G’s unified syntax for several data types eliminates the need to have multiple files in different formats by acting as a container including all the information from legacy formatted files. This feature is significantly different from other formats. MPEG-G is not just another optimized SAM or FASTQ compressor. Still, it provides a single syntax to represent raw and aligned data, genome assemblies, and very soon variants and annotations. As well, indexing is embedded in the compressed data.
Another critical attribute is the open specification. The MPEG-G standard is entirely specified as an open document and related open-source reference software. Therefore, any organization can implement it, avoiding the risk of being locked in closed proprietary solutions.
What good can a unified single syntax do?
MPEG-G approach starts from a fundamental idea: the abstraction between the actual genomic information (e.g., sequences, read names, quality values, alignment information) and the way how it is represented (the formats). The direct effect of this approach is that the entire information represented by legacy formats, such as FASTA, FASTQ, BAM, SAM, CRAM, and soon VCF, can be equally represented by the MPEG-G format. This means that any analytical tool can either work directly on the MPEG-G format or continue working on legacy formats through fully lossless transcoding, still preserving full compatibility. This aspect is particularly important when talking about data sharing.
So, the compatibility between MPEG-G and legacy formats is a cornerstone for interoperability amongst genomic data. For example, any research center that uses MPEG-G formatted genomic data files – a single file including the information from FASTA, FASTQ, SAM, BAM, and/or CRAM – can easily share genomic data with other research facilities minimizing the number of files shared.
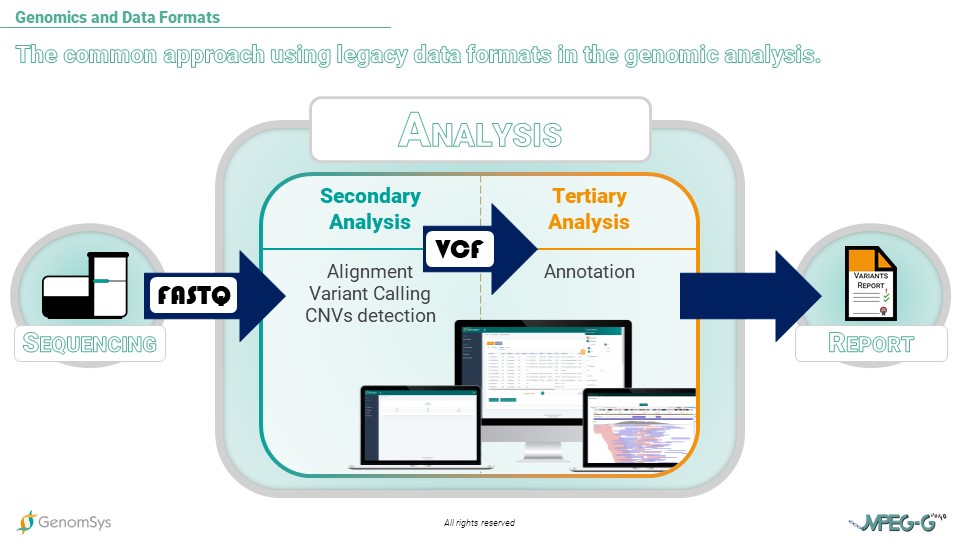
Graphic 1 – Workflow of genomic testing, including the common data formats during Sequencing, Analysis, and Report.
For example, if a research group in Switzerland wants to share 100 Whole-Exome Sequencing (WES) datasets with 100x mean coverage with another research group in Spain, in legacy formats, the data exchange would include for each sample a pair of FASTQ gzipped files as the raw genomic data.
The number of files for the 100 WES samples would be 200 single files. MPEG-G, thanks to its hierarchical structure based on datasets and dataset groups, allows many options in terms of aggregation of files, from, e.g., sharing only 100 individual files (one per sample) to sharing a single MPEG-G file including all the 100 samples, where each sample can be represented as a different dataset group; in both cases, the MPEG-G file(s) would include the same information as the 200 files for legacy formats. The data size would decrease from roughly 1’148 GB of data for the legacy formatted files (FASTQ gzipped pairs) to 473 GB of MPEG-G formatted data.
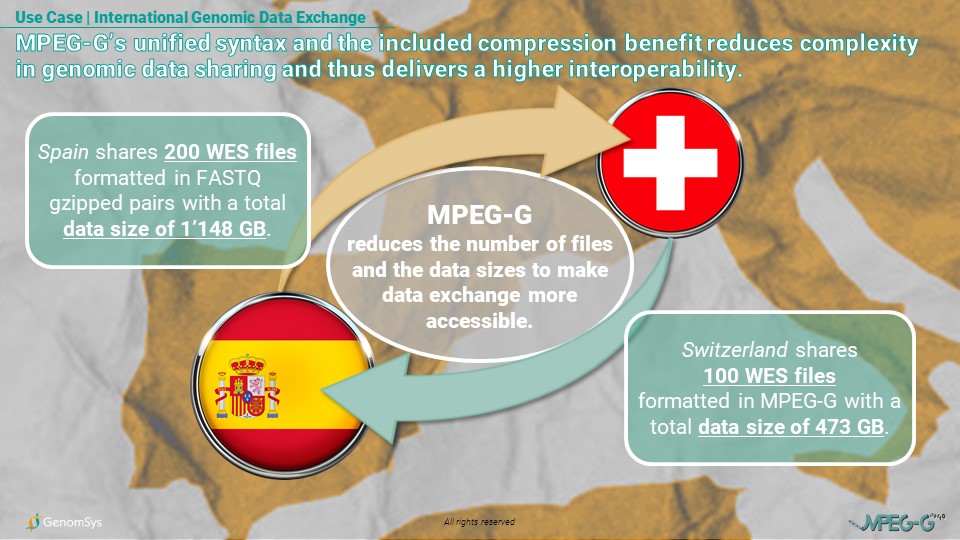
Graphic 2 – Use Case for the interoperability of the MPEG-G format compared to legacy formats concerning international data exchange amongst research groups in Switzerland and Spain.
So, this unified syntax and the included compression benefit from MPEG-G allows a reduction of complexity. For this specific example, the single syntax cuts the number of files in half. MPEG-G’s compression shrinks the data sizes; thus, it simplifies the data exchange because fewer and smaller files can be sent, increasing the interoperability between the two research groups.
The ISO standard is open, but what does that mean for laboratories?
The openness of the MPEG-G format, due to the nature of any ISO working group in developing standards, helps to establish a certain degree of conformity and thus interoperability. A specific part of the standard is dedicated to conformity: part 5. In this part, highly detailed conformity tests are defined to cover every single feature/bit defined in the standard. Conformity is enforced at both the level of the bitstream to provide a way to validate the output of any conformant encoder and the decoding process so that every single part of the decoding process can be validated. One can claim his codec to be conformant only once it has passed all conformity tests: this is essential, and it guarantees that all MPEG-G codecs around will speak the same language, i.e., they are interoperable. There is no room for ambiguities in this process, as ambiguities would break interoperability. This notably aims at any software developing company in genomics to freely adapt MPEG-G into any software solution for genomics they want to build, including the possibility of still interacting with legacy formats.
However, at first sight, the openness of the ISO standard is only a direct benefit for genomic software companies, but laboratories are also indirectly affected by this circumstance. Using an ISO standard in an open way leads to a wider variety of highly interoperable genomic software due to the MPEG-G format. Another effect is that by using an ISO standard, the certification of the software becomes more uncomplicated and faster. Thus, the laboratories can choose from a variety of certified software that is in itself highly interoperable.
For the genomic software developing company, the backward compatible lossless transcoding – an effect of its built-in openness – is meaningful and allows future further development of the software based on MPEG-G to still be compatible with previous software-based MPEG-G. Current problems due to different versions of the legacy formats can cause a decrease in interoperability. At the same time, MPEG-G was developed to avoid these issues and provide a “fallback” if needed, i.e., full backward compatibility among possible different versions/updates of the standard. This circumstance then directly affects researchers and laboratories using different software versions and still being able to use it once their own version is outdated.
As we talk about genomic data formats and thus a part of a medical device, certification, and accreditation are important. It is worth mentioning, being the only ISO standard for the representation of genomic data, that the accompanied conformity directly impacts the certification of medical devices and ultimately for the laboratory, which will be allowed to only use certified tools in their analysis after the new IVDR. So by using MPEG-G, software developing companies can simplify their certification process, as they are using a globally recognized ISO format, and provide laboratories with a reliable solution supported by ISO, which then helps the laboratory in their own accreditation process.
So, MPEG-G’s standardized open format, coming from the ISO institution, delivers the needed interoperability for the entire genomics environment, including the software developing companies and genomic laboratories – another way to look at interoperability amongst the genomic field in itself.
GenomSys professional’s solution leveraging the interoperability feature from MPEG-G
The CE marked GenomSys Variant Analyzer is natively running on the MPEG-G format and allows converting FASTQ data seamlessly, coming directly from the sequencing machine, into MPEG-G before each analysis. It is the worldwide first genomic software that uses the MPEG-G format to enable laboratories and researchers worldwide to leverage the high interoperability feature of the ISO format.
By using GenomSys Variant Analyzer, the entire analysis pipeline, including steps working on aligned data, can take advantage of all its features, from compression to Selective Access, thus minimizing the overall processing time and the storage and memory footprints as well as its single syntax for an easier way to share data with research colleagues.
Please find our Fact-Card on MPEG-G under the link with information on the Interoperability feature and a specific use-case.
Stay tuned for our next article in the series “MPEG-G, can I eat it?” on the benefits of the MPEG-G’s built-in security highly important when talking about genomic data and our most precious information, our DNA, next week.
By Lucas Laner and Daniele Renzi on March 23rd, 2022.
[1] McGuire, A.L., Gabriel, S., Tishkoff, S.A. et al. The road ahead in genetics and genomics. Nat Rev Genet 21, 581–596 (2020). https://doi.org/10.1038/s41576-020-0272-6
[2] Bernasconi, Anna, and Stefano Ceri. 2022. “Interoperability of COVID-19 Clinical Phenotype Data with Host and Viral Genetics Data” BioMed 2, no. 1: 69-81. https://doi.org/10.3390/biomed2010007
[3] Baha Zeidan. COVID-19 Highlights the Need for Interoperability in Healthcare (2021). https://www.bio-itworld.com/news/2021/04/16/covid-19-highlights-the-need-for-interoperability-in-healthcare


 / pixabay
/ pixabay

