Every second, physicians, nurses, pharmacists, researchers, and health regulators worldwide generate and use large numbers of essential healthcare information critical to their work: saving lives. Any decision in medicine is based on scientific data, be it for prevention or treatment. Today with all the technological breakthroughs over the past decades, patient care is generating a vast pool of data. The use and exchange of this data have been crucial for new discoveries that lead to a deeper understanding of how our body works and, thus, how to prevent or treat illnesses.
In this context, to better understand the amount of data in healthcare, about 30% of all data today is generated in healthcare.[1] By 2025, its growth rate is expected to reach 36% and thus growing faster than manufacturing, financial services and media & entertainment.[2] For the average patient, this means annually, 80 MB of new healthcare data is generated today and by 2025 close to 110 MB, which would mean for the population in Europe a generation of 82’104 TB of personal data each year.[3][4]
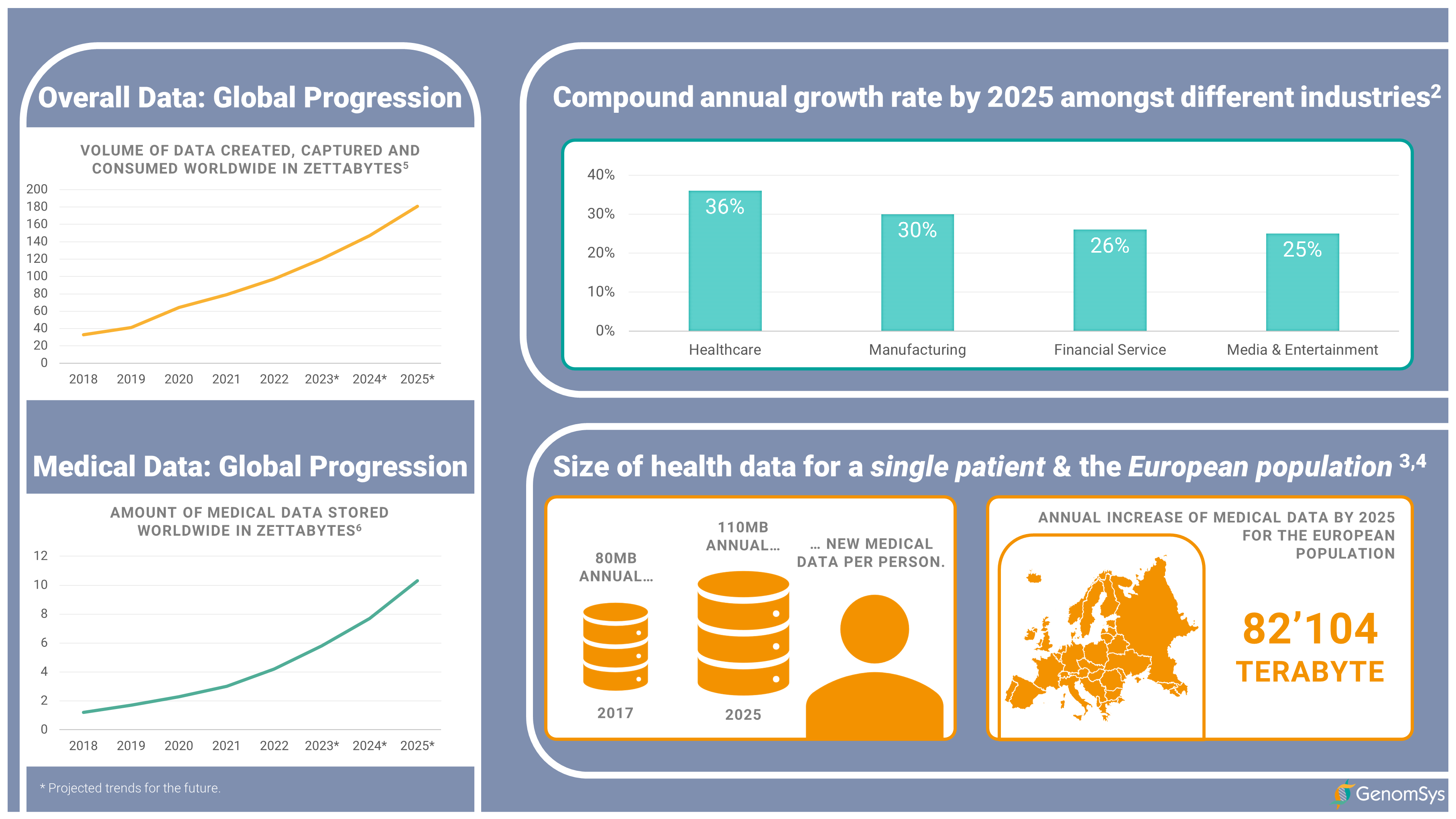 Figure 1 – Global progression of overall data and medical data, the compound annual growth rate by 2025 for different industries and size of health data for a single patient and the European population.[2][3][4][5][6]
Figure 1 – Global progression of overall data and medical data, the compound annual growth rate by 2025 for different industries and size of health data for a single patient and the European population.[2][3][4][5][6]
How vital health data is shown, for example, by the recent initiative of the European Commission called European Health Data Space (EHDS), particularly the storage, reuse and exchange of up-to-date data.
“Health data are the blood running through the veins of our healthcare systems. The COVID-19 pandemic has shown that up-to-date health data are essential to take well-informed public health measures and respond to crises. The pandemic has also triggered a considerable acceleration in the uptake of digital tools. Unfortunately, there are still complex obstacles that make it difficult to reach the full potential of digital health and health data.
The European Health Data Space will overcome these obstacles. It is a health-specific data sharing framework establishing clear rules, common standards and practices, infrastructures and a governance framework for using electronic health data by patients and for research, innovation, policy making, patient safety, statistics or regulatory purposes.” [7]
But data generation is solely a starting point in times of sustainability and the need for faster turnaround times of diagnosis. Reusing the former generated information and establishing meaningful connections is vital for us to support more efficient research, accelerate more convenient patient care processes, and economically improve our healthcare system.
One of the drivers for this aggregation of data is the medical field of genetics. Over the past 20 years, genomics – a relatively young discipline compared to others in medicine – with connections to all other areas of medicine, as our DNA is responsible for all of our processes within our bodies, has evolved through the Next-Generation Sequencing (NGS) technology to a powerful generator of medical data. We are witnessing how the recent technological breakthroughs have empowered the scaling-up of genomic applications, as it is now possible to sequence the whole genome cost-effectively.
This large amount of information can give insights into diseases in various fields of medicine, as it has already improved the treatment of cancer patients or diagnostics of rare pediatric diseases. Still, there is much more to discover on how our genes affect our body and how these insights can lead to other applications within patient care. But it is a question of time and research effort until correlations between variations in our DNA and illnesses are discovered, which then can function as starting points for treatments or preventive measures.
Considering broader scope genomic applications like the sequencing of the whole-coding region (Whole-Exome Sequencing, i.e., WES) or the whole-genome (Whole-Genome Sequencing, i.e., WGS), the concept of data reusability gains more relevance.
Next-generation sequencing (NGS) defines massively parallel sequencing technology that in the last decades has revolutionized genomic research. NGS provides high-throughput and scalable methods supporting a wide set of applications in research and diagnostics.
Whole-exome sequencing (WES) is a technique for analyzing all the protein-coding regions in the genome (i.e., exome).
Whole-genome sequencing (WGS) is a sequencing technique for analyzing the entire genome.
A common approach for genetic testing today is gene panels. Gene panels and their limited region target – usually ranging from 5-200 genes – do not provide a base for data reuse, as they can only answer a limited number of medical questions. But considering the recent trend of steady shifting toward WES and, in some places, WGS as an initial diagnostic tier, data reuse can be in place to fully deliver its benefits.
The reuse of data can have an impact on various stakeholders in healthcare. In the following, we would like to explain further the benefits of reusing genomic data, from WES or WGS, for patients, the healthcare system itself and health insurance.
Patient care
The genetic testing process requires patients to undergo genetic counseling and the actual drawing of the blood or saliva for DNA extraction. After the DNA has been sequenced and the data analyzed, the patients get informed about the results. Once an analysis is performed, the data generated are seldom used for other purposes, e.g., research or additional compatible analysis.
Patients can benefit directly from reusing their health data in terms of time-saving. Individuals need to spend time providing samples on which the data is generated. This process regarding genomic data includes the consultation of geneticists. During this, the patient receives essential information about the procedure of genetic testing, what the analysis can and cannot determine, the impact of the result for themselves and their families, and the actual drawing of the blood or saliva for the DNA extraction. Reusing the initially generated data, especially from whole-genome analysis, allows the patient to save time by skipping some of the beforementioned steps during the genetic testing.
Especially looking at the expansion of personal genetic testing, such as pharmacogenetics or nutrigenetics, the reuse of WES or WGS data could enable timely analysis based on new findings and make sample collection as well as sequencing a one-time effort.
Healthcare system
As mentioned earlier, most genetic testing today is based on sequencing a few genes in gene panels. But regarding WES and WGS and their potential broader application in the future due to cost droppings in sequencing, reusing genomic data could benefit our healthcare system.
By reusing genomic or medical data, the healthcare system can leverage the formerly generated data and use it to learn from it. This idea is termed “the learning health system,” and it implies that the reuse of the data collected as a by-product of clinical care to improve the performance of our health care system and provide individual patients with the best possible information about their diagnostic and treatment choices.[8]
Insurance
When discussing health data, a particular term has been floating around for quite some time now: Electronic Health Records (EHR). EHRs collect patients’ health data in a digital format. This digitization of patient-centered records can be updated in real-time and made available instantly and securely to any authorized recipient. Besides this direct benefit for the patient, as he can share vital information amongst his multiple physicians at any time, electronic health record systems are built to gain further an entire picture of the patient to personalize the care and ultimately improve the success of any treatment.[9]
This entire picture can also allow the business side of medicine, an estimated multi-trillion-dollar industry, to become more efficient, particularly in health insurance. Insurance companies can use their claims databases to perform actuarial calculations that estimate risk for patient populations so they can set realistic insurance premiums.[8]
The reuse of data seems to be overall beneficial to patients, researchers and the entire healthcare system but there are still barriers that prohibit the full reuse of data.
Storage costs of vast amount of data
Over the past 10 years, genetics has generated a vast amount of data due to the rise of NGS and its high-throughput sequencing in germline and oncology testing. Just to put the volume a little bit into perspective, a single human genome sequence alone takes up 200GB of storage, and by 2025, the worldwide storage space would grow to an estimate of 40 exabytes (1 exabyte = 1’073’741’824 gigabyte) or as the National Human Genome Research Institute puts it: “In comparison, five exabytes could store all of the words ever spoken by human beings.”[10]
These amounts of data need to be stored and potentially stored for a lifetime. Currently, traditional storage media is pushed to the limits of its capacity. Laboratories no longer have the space to store the mass of data locally. The solution is the cloud. And it has become more and more critical amongst most industries as data storage is being outsourced to cloud service providers in the effort to save costs.[11]
Cloud-based storage provides several advantages, such as low per-GB prices, scalability, and minimal fixed costs; however, while these solutions advertise seemingly simple usage-based pricing plans, practical cost analysis of cloud storage for NGS data storage is not straightforward.[12]
Although cloud-based storage can keep costs down, two factors still need to be considered. Firstly, fewer data should be generated, which can be achieved by a well-established process of data reuse, and secondly, genetic data should be stored as efficiently as possible in a format that leaves out redundant information and thus generates less data.[12]
Storage of health data in silos
Another problem healthcare faces today is the storage of health data in silos, limiting the exchange and reuse. As the data is generated in different locations with different formats, it is still common for patient information to be incomplete at the point of care. This disconnection causes multiple medical records and everything from disease information to full medication lists to create a web of duplicates and redundant information, making decisions in the health care process longer and more complex than they have to be.[13]
The reason behind the silos is the missing standard for health data and, therefore, the lack of interoperability. But this lack of standardization is the common regret for anything new. Today, there is a multitude of data-sharing initiatives with the ambition to establish universal interoperability, which is a huge undertaking. But the transformation to standardized formats – ultimately one format for full interoperability – will need time. Experts estimate that standardizing health data might take up to 10 years, but the benefits vastly outweigh the efforts.
In genetics, a standardized format could become necessary, which was already a great help in a completely different industry (audio & video) based on large data sets. This is the MPEG-G standard for the representation of genetic data developed by an ISO working group. The structure for this standard aims to provide interoperability due to its unified syntax that can represent all genetic data and metadata. Usually, genetic information is stored within multiple legacy formats with different features, requiring conversion steps from one another, which means losing valuable time and carries the risk that all the various files are not stored in one place. The newly developed ISO standard MPEG-G can represent genetic data in a single file that includes all the related information, more efficiently compressed, and thus ready to break the silo storage of connected genetic health information gathered in one place.
Legal barriers in terms of privacy
The European academy network formed a working group to address current challenges that hinder a more efficient way to share and reuse health data and ensure effective collaboration with public research institutions. In their report International Sharing of Personal Health Data for Research published in April 2021, they demand easier sharing of pseudonymized health data with researchers worldwide, under article 46 of the General Data Protection Regulation (GDPR), overcoming European borders.[14]
Although the technology for sharing pseudonymized health data would be there, which allows a rapid exchange of data amongst research groups, dealing with data privacy and its accompanying legislation slows down the entire process. According to the report, data transfer challenges arise from the legal conflict between the European Union and other countries’ legislation since it is not possible to sign contracts outside the EU – being subject to the GDPR – because of missing viable legal mechanism for sharing data for public sector research.[14]
This leads to a slowdown in medical progress and does not exploit the full potential of research, ultimately affecting the quality of healthcare and thus weakening the population’s health.
Particularly for genetics and its different laws amongst the EU, this legislation patchwork is a barrier to the appropriate reuse of genetic data. It adds complexity – basically slows down the process of helping patients – to an already complex field. For example, French national laws allow tests to be covered under biomedical regulation, while Germany and Austria have laws specific to genetic testing. Other countries don’t have specific laws but regulate them as health services.[14]
But hope is on the horizon with, for example, the SIENNA project. This EU-funded effort will release guidelines for harmonized ethical frameworks and codes of conduct in the genetic field, as well as address issues of the inconsistently regulated field of genetic Direct-to-Consumer (DtC) tests.1[14]
Although there are still barriers that hinder the full adaption of data reuse, the need to leverage the reuse of health data, especially genetic data with its vast size, will be the future. It aligns with the common trend of our time to become more sustainable and leverage synergies to become more efficient in healthcare.
What role does GenomSys play here
We at GenomSys want to play a particular role in this. Our solutions based on the MPEG-G standard fulfill the promise to make genetic testing more efficient and establish a truly personalized medicine.
Our Ecosystem developed on the idea of supporting the professionals with more efficient applications (GenomSys Variant Analyzer and GenomSys MPEG-G Toolkit) for their routine work and enabling individuals to conveniently and actively participate in the process of genetic testing (GenomYou).
GenomSys Variant Analyzer and GenomSys MPEG-G Toolkit are developed to provide geneticists with solutions that result in less time spent on the analysis, smaller file sizes and a new way to facilitate the combination of multiple genetic data and metadata into a single file that is interoperable with any other format. We foresee that these features are, to a certain degree, already needed and will become more critical for genetics in the future in the quest for efficient genetic testing and to connect all health data for an improved way to help patients worldwide.

“Every day, new information helps us broaden our comprehension of genomic variation. Sharing this information can maximize our knowledge and lead us to shift toward a truly personalized perspective in healthcare,”
says Luca Trotta, Chief Scientific Officer @ GenomSys.
The aspect for individuals of our Ecosystem is the vision of empowering patients to leverage the most personal tool in their hand – the smartphone – to be more engaged in managing their health and get truly personalized medicine through genetics. Our smartphone has become the ultimate gateway for most interactions in our daily lives. Be it for finding directions, connecting with our friends and family or gathering information. In our opinion, genetics, due to its nature as each of our DNA is unique, is prone to be a key accelerator of personalized medicine. This field, paired with our most used tool today – the smartphone – allows patients conveniently to become an active part in taking care of their health and interacting with professionals in genetics and healthcare providers in the complex field of genetics.
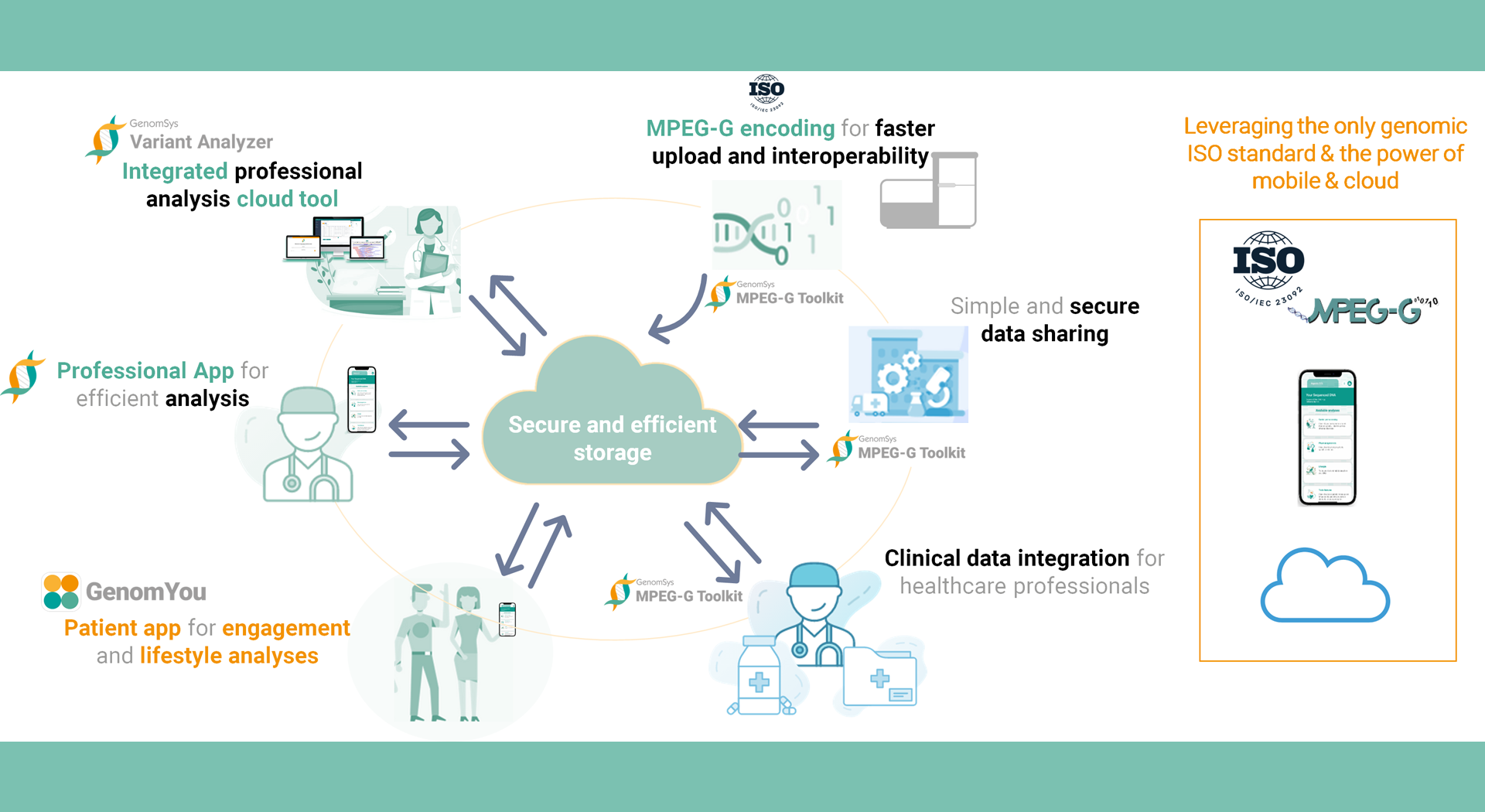 Figure 2 – GenomSys EcoSystem. Connecting professionals and individuals in genetic testing more efficiently and conveniently for a future of true personalized medicine.
Figure 2 – GenomSys EcoSystem. Connecting professionals and individuals in genetic testing more efficiently and conveniently for a future of true personalized medicine.
By Lucas Laner on July 13th, 2022.
References:
[1] RBC Capital Markets; The healthcare data explosion (2018). https://www.rbccm.com/en/gib/healthcare/episode/the_healthcare_data_explosion
[2] David Reinsel, John Gantz, John Rydning; The Digitization of the World From Edge to Core (2018). https://www.seagate.com/files/www-content/our-story/trends/files/idc-seagate-dataage-whitepaper.pdf
[3] Nick Culbertson; The Skyrocketing Volume Of Healthcare Data Makes Privacy Imperative (2021). https://www.forbes.com/sites/forbestechcouncil/2021/08/06/the-skyrocketing-volume-of-healthcare-data-makes-privacy-imperative
[4] Deutsche Stiftung Weltbevölkerung; Soziale und demografische Daten weltweit DSW-DATENREPORT (2019). https://www.dsw.org/wp-content/uploads/2019/12/DSW-Datenreport-2019.pdf Number of inhabitants in Europe 746.4 million. The countries included in Europe: Albania, Andorra, Austria, Belarus, Belgium, Bosnia and Herzegovina, Bulgaria, Channel Islands, Croatia, Czech Republic, Denmark, Estonia, Finland, France, Germany, Greece, Hungary, Iceland, Ireland, Italy, Kosovo, Latvia, Liechtenstein, Lithuania, Luxembourg, Malta, Marino, Monaco, Montenegro, Netherlands, North Macedonia, Norway, Poland, Portugal, Republic of Moldova, Romania, Russia, San Marino, Serbia, Slovakia, Slovenia, Spain, Sweden, Switzerland, Ukraine and United Kingdom.
[5] Statista Research Department; Volume of data/information created, captured, copied, and consumed worldwide from 2010 to 2025 (2021). https://www.statista.com/statistics/871513/worldwide-data-created/
[6] Dr. Hans Christian Müller; Gesundheitsdaten – Ausgangspunkt für eine lernende Gesundheitsversorgung (2021). https://www.handelsblatt.com/downloads/27790440/1/gesundheitsdaten.pdf
[7] European Comission; Questions and answers – EU Health: European Health Data Space (EHDS) (2022). https://ec.europa.eu/commission/presscorner/detail/en/QANDA_22_2712
[8]C. Safran; Update on Data Reuse in Health Care (2017). https://www.thieme-connect.com/products/ejournals/html/10.15265/IY-2017-013
[9] HealthIT.gov; What is an electronic health record (EHR)? (2019) https://www.healthit.gov/faq/what-electronic-health-record-ehr
[10] National Human Genome Research Institute; Genomic Data Science (2022). https://www.genome.gov/about-genomics/fact-sheets/Genomic-Data-Science
[11] Matthias Janson; 2020 überholt die Cloud lokale Speichermedien (2019). https://de.statista.com/infografik/18231/cloud-vs-lokaler-speicher/
[12] Niklas Krumm, Noah Hoffman, Practical estimation of cloud storage costs for clinical genomic data, Practical Laboratory Medicine, Volume 21, 2020, e00168, ISSN 2352-5517, https://doi.org/10.1016/j.plabm.2020.e00168.
[13] Michael Schroeder; How hospitals are breaking down data silos to improve patient care (2022). https://medcitynews.com/2022/05/how-hospitals-are-breaking-down-data-silos-to-improve-patient-care/
[14] Cheng A, Guzman CEV, Duffield TC, Hofkamp H. Advancing Telemedicine Within Family Medicine’s Core Values. Telemed J E Health. 2021 Feb;27(2):121-123. doi: 10.1089/tmj.2020.0282. Epub 2020 Jul 28. PMID: 32744897; PMCID: PMC7888289.





