Genetics is a vibrant, relatively new field of medicine; thanks to the implementation of massive parallel sequencing with next-generation sequencing (NGS) over the last 10 years, many new applications in genomics have appeared. With this article, we want to shine a light on two particular trends for the genetic diagnostics field: the evolution from the gene-panel approach to the whole-exome and explain a new application area for genetic testing through liquid biopsy.
The ever-increasing adoption of NGS-based methods in clinical settings has dramatically advanced the identification of the genetic causes of Mendelian phenotypes, providing support for diagnostic, preventive, and therapeutic strategies, and supporting the development of personalized medicinal approaches is expected in upcoming years [1, 2]. As described in one of our previous articles, “Genetic analysis how is it currently done?” it is now economically possible to sequence the entire genome within a couple of days [3].
Evolution from gene panels to whole-exome diagnostics but not yet whole-genome.
The trend of sequencing more regions on the DNA is expected to spill over in other areas besides rare diseases. This leaves the geneticist with more options to examine, with new findings arising every day and ultimately leading to a higher quality of reports.
Additionally, the trend to initially sequence larger areas can also be observed from an economic perspective. According to a market intelligence report by ResearchAndMarkets.com, the global market for whole-genome and whole-exome sequencing is expected to increase more than ten times by 2029 [4].
For the clinical use of NGS, it is crucial to select the most appropriate testing applications for the specific diagnostic requirements. Gene-panels (targeted sequencing), whole-genome sequencing (WGS), and whole-exome sequencing (WES) are among the most widely adopted methods in clinical genetics. For a deeper explanation of each of these methods, see our overview by our Chief Scientific Officer Luca: NGS-based methods and rare diseases.
Over the last couple of years, the WES method is increasingly preferred for routine diagnostics, especially for rare diseases where almost 85% of the disease-causing variants lie within the coding region. WES more and more replaces Gene-panels due to their limitation in expanding the analysis over the specified targets. The complete extension to a WGS approach is still in its early stages but still limited in for routinely application diagnostics. The reasons for this are the prohibitively high costs for most researchers – almost twice than WES – while the diagnostic utility of sequencing the whole-genome did not show significant differences in monogenic diseases compared to the whole-exome approach [5].
Liquid biopsy – transforming genetic analysis to a routine test in the prevention of cancer
Today genetic testing is limited to only determine health risks based on the inherited DNA for each individual. Although in some instances DNA can change at the somatic level and can cause diseases, for example, tumors, current genetic analysis from a saliva or blood sample cannot show these changes.
Liquid biopsy represents a new application possibility, examining the low concentration of cell-free tumor DNA within the bloodstream to detect possible tumor development. The approach is similar to the existing analysis of a biopsy sample from tumor tissue. Still, it occurs earlier while tumor growth is at an early stage, and it is definitely less invasive for the probands [6].
For Liquid biopsy, the same sequencing technique – NGS – is applied for the genetic analysis of germline DNA, although the target of interest is the cell-free tumor DNA instead of the DNA from “healthy” cells. Based on highly complex bioinformatic steps during the analysis, it is now possible to detect minimal DNA concentrations of the cell-free tumor DNA and map them to tumor references.
This evolution of analysis of non-germline DNA can have substantial implications for how cancer is prevented, treated, and even monitored after the treatment. Aside from the benefits for the patients, early cancer detection increases the chance of survival; this method can provide researchers active surveillance on the molecular changes in a tumor [6].
Experts claim that, through this feature of liquid biopsy, if evidence from further clinical trials confirms the higher utility, an annual blood test for individuals at increased risk of tumors may be another helpful tool[7].
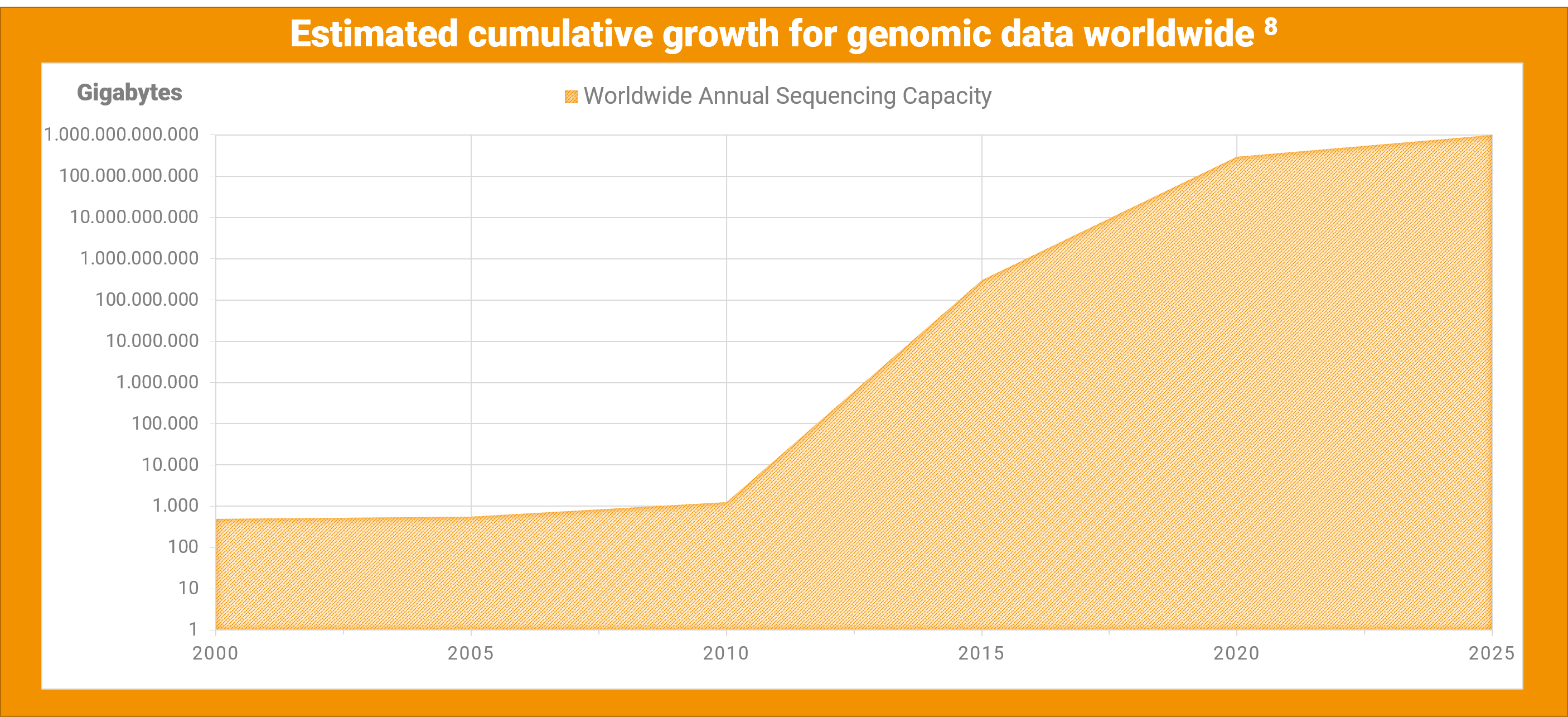
Both of these trends foreshadow an increase in genomic data. Based on the historical growth rate for sequenced genomes, estimations are that by the year 2025, we will succeed 1,000,000 terabytes of genomic data worldwide[8]. With the increasing amount of data, there will be more need for more efficient data storage solutions, resulting in lower storage costs and more energy efficiency.
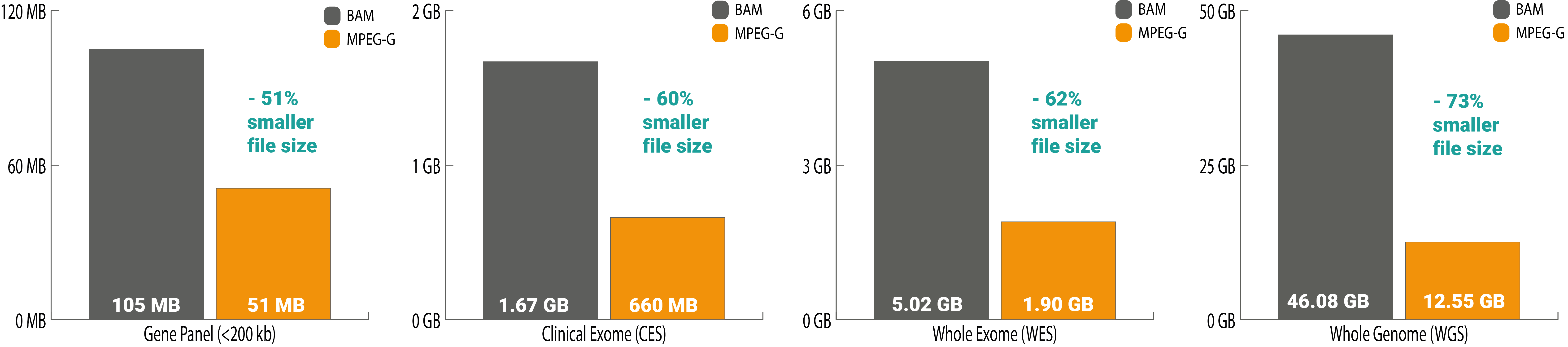
As for managing any electronic data, addressing the storage for such an amount of genetic data is essential. With the high compression benefit of the ISO/23092 genomic data format (MPEG-G), a whole-exome file’s data size is reduced by more than 60%. Lowering storage costs, as well as decreasing the energy uptake to store this data.
By Lucas Laner on August 25, 2021.
References:
[1] Rabbani B, Tekin M, Mahdieh N. The promise of whole-exome sequencing in medical genetics. J Hum Genet 2014 Jan;59(1):5-15.[2] Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD, et al. The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am J Hum Genet 2015 Aug 6;97(2):199-215.
[3] https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Fact-Sheet
[4] https://www.globenewswire.com/news-release/2020/06/02/2042370/0/en/Global-Whole-Genome-and-Exome-Sequencing-Market-2019-to-2029-Focus-on-Product-Workflow-Application-End-user-Country-Data-and-Competitive-Landscape.html
[5] Clark, M.M., Stark, Z., Farnaes, L. et al. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. npj Genomic Med 3, 16 (2018). https://doi.org/10.1038
[6] https://www.cancer.gov/publications/dictionaries/cancer-terms/def/liquid-biopsy
[7] https://www.nature.com/articles/d41586-020-00844-5
[8] Stephens et al. (2015). Big Data: Astronomical or Genomical?. PLoS biology. 13. e1002195. 10.1371/journal.pbio.1002195.
Pictures source:
lovini| pixabay